Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1
- PMID: 23429703
- PMCID: PMC3753690
- DOI: 10.1126/science.1228792
Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1
Abstract
Cellular growth signals stimulate anabolic processes. The mechanistic target of rapamycin complex 1 (mTORC1) is a protein kinase that senses growth signals to regulate anabolic growth and proliferation. Activation of mTORC1 led to the acute stimulation of metabolic flux through the de novo pyrimidine synthesis pathway. mTORC1 signaling posttranslationally regulated this metabolic pathway via its downstream target ribosomal protein S6 kinase 1 (S6K1), which directly phosphorylates S1859 on CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase), the enzyme that catalyzes the first three steps of de novo pyrimidine synthesis. Growth signaling through mTORC1 thus stimulates the production of new nucleotides to accommodate an increase in RNA and DNA synthesis needed for ribosome biogenesis and anabolic growth.
Conflict of interest statement
All authors have reviewed the manuscript and declare no competing financial interests.
Figures
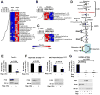

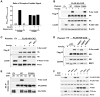

Comment in
-
mTORC1 signaling aids in CADalyzing pyrimidine biosynthesis.Cell Metab. 2013 May 7;17(5):633-5. doi: 10.1016/j.cmet.2013.04.018. Cell Metab. 2013. PMID: 23663734
References
-
- Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans. 2012 Feb;40:168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

