Molecular pathways: extracting medical knowledge from high-throughput genomic data
- PMID: 23430023
- PMCID: PMC3686839
- DOI: 10.1158/1078-0432.CCR-12-2093
Molecular pathways: extracting medical knowledge from high-throughput genomic data
Abstract
High-throughput genomic data that measures RNA expression, DNA copy number, mutation status, and protein levels provide us with insights into the molecular pathway structure of cancer. Genomic lesions (amplifications, deletions, mutations) and epigenetic modifications disrupt biochemical cellular pathways. Although the number of possible lesions is vast, different genomic alterations may result in concordant expression and pathway activities, producing common tumor subtypes that share similar phenotypic outcomes. How can these data be translated into medical knowledge that provides prognostic and predictive information? First-generation mRNA expression signatures such as Genomic Health's Oncotype DX already provide prognostic information, but do not provide therapeutic guidance beyond the current standard of care, which is often inadequate in high-risk patients. Rather than building molecular signatures based on gene expression levels, evidence is growing that signatures based on higher-level quantities such as from genetic pathways may provide important prognostic and diagnostic cues. We provide examples of how activities for molecular entities can be predicted from pathway analysis and how the composite of all such activities, referred to here as the "activitome," helps connect genomic events to clinical factors to predict the drivers of poor outcome.
Figures
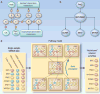
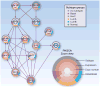
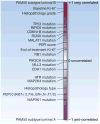
References
-
- Kschischang, Frey, Loeliger Factor graphs and the sum-product algorithm. IEEE Transactions on Information Theory. 2001:47.
-
- Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. Journal of computational biology: a journal of computational molecular cell biology. 2011;18:507–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

