Novel tryptophan metabolism by a potential gene cluster that is widely distributed among actinomycetes
- PMID: 23430264
- PMCID: PMC3617294
- DOI: 10.1074/jbc.M112.436451
Novel tryptophan metabolism by a potential gene cluster that is widely distributed among actinomycetes
Abstract
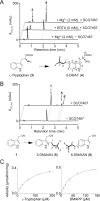
The characterization of potential gene clusters is a promising strategy for the identification of novel natural products and the expansion of structural diversity. However, there are often difficulties in identifying potential metabolites because their biosynthetic genes are either silenced or expressed only at a low level. Here, we report the identification of a novel metabolite that is synthesized by a potential gene cluster containing an indole prenyltransferase gene (SCO7467) and a flavin-dependent monooxygenase (FMO) gene (SCO7468), which were mined from the genome of Streptomyces coelicolor A3(2). We introduced these two genes into the closely related Streptomyces lividans TK23 and analyzed the culture broths of the transformants. This process allowed us to identify a novel metabolite, 5-dimethylallylindole-3-acetonitrile (5-DMAIAN) that was overproduced in the transformant. Biochemical characterization of the recombinant SCO7467 and SCO7468 demonstrated the novel L-tryptophan metabolism leading to 5-DMAIAN. SCO7467 catalyzes the prenylation of L-tryptophan to form 5-dimethylallyl-L-tryptophan (5-DMAT). This enzyme is the first actinomycetes prenyltransferase known to catalyze the addition of a dimethylallyl group to the C-5 of tryptophan. SCO7468 then catalyzes the conversion of 5-DMAT into 5-dimethylallylindole-3-acetaldoxime (5-DMAIAOx). An aldoxime-forming reaction catalyzed by the FMO enzyme was also identified for the first time in this study. Finally, dehydration of 5-DMAIAOx presumably occurs to yield 5-DMAIAN. This study provides insight into the biosynthesis of prenylated indoles that have been purified from actinomycetes.
Figures






References
-
- Newman D. J., Cragg G. M. (2007) Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477 - PubMed
-
- Clardy J., Walsh C. (2004) Lessons from natural molecules. Nature 432, 829–837 - PubMed
-
- Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H., Shiba T., Sakaki Y., Hattori M., Omura S. (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 - PubMed
-
- Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C. H., Kieser T., Larke L., Murphy L., Oliver K., O'Neil S., Rabbinowitsch E., Rajandream M. A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

