Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation
- PMID: 23431030
- PMCID: PMC3605469
- DOI: 10.1101/gad.207654.112
Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation
Abstract
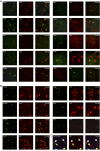
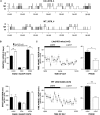
Hypocretin (orexin; Hcrt)-containing neurons of the hypothalamus are essential for the normal regulation of sleep and wake behaviors and have been implicated in feeding, anxiety, depression, and reward. The absence of these neurons causes narcolepsy in humans and model organisms. However, little is known about the molecular phenotype of these cells; previous attempts at comprehensive profiling had only limited sensitivity or were inaccurate. We generated a Hcrt translating ribosome affinity purification (bacTRAP) line for comprehensive translational profiling of all ribosome-bound transcripts in these neurons in vivo. From this profile, we identified >6000 transcripts detectably expressed above background and 188 transcripts that are highly enriched in these neurons, including all known markers of the cells. Blinded analysis of in situ hybridization databases suggests that ~60% of these are expressed in a Hcrt marker-like pattern. Fifteen of these were confirmed with double labeling and microscopy, including the transcription factor Lhx9. Ablation of this gene results in a >30% loss specifically of Hcrt neurons, without a general disruption of hypothalamic development. Polysomnography and activity monitoring revealed a profound hypersomnolence in these mice. These data provide an in-depth and accurate profile of Hcrt neuron gene expression and suggest that Lhx9 may be important for specification or survival of a subset of these cells.
Figures



Similar articles
-
Evolutionarily conserved regulation of hypocretin neuron specification by Lhx9.Development. 2015 Mar 15;142(6):1113-24. doi: 10.1242/dev.117424. Epub 2015 Feb 27. Development. 2015. PMID: 25725064 Free PMC article.
-
Neural substrates of awakening probed with optogenetic control of hypocretin neurons.Nature. 2007 Nov 15;450(7168):420-4. doi: 10.1038/nature06310. Epub 2007 Oct 17. Nature. 2007. PMID: 17943086 Free PMC article.
-
Transgenic Archaerhodopsin-3 Expression in Hypocretin/Orexin Neurons Engenders Cellular Dysfunction and Features of Type 2 Narcolepsy.J Neurosci. 2019 Nov 20;39(47):9435-9452. doi: 10.1523/JNEUROSCI.0311-19.2019. Epub 2019 Oct 18. J Neurosci. 2019. PMID: 31628177 Free PMC article.
-
The role of hypocretins (orexins) in sleep regulation and narcolepsy.Annu Rev Neurosci. 2002;25:283-313. doi: 10.1146/annurev.neuro.25.112701.142826. Epub 2002 Mar 20. Annu Rev Neurosci. 2002. PMID: 12052911 Review.
-
The Hypocretin/Orexin Neuronal Networks in Zebrafish.Curr Top Behav Neurosci. 2017;33:75-92. doi: 10.1007/7854_2016_59. Curr Top Behav Neurosci. 2017. PMID: 28012092 Review.
Cited by
-
Potential therapeutic effect of NK1R antagonist in diabetic non-healing wound and depression.Front Endocrinol (Lausanne). 2023 Jan 4;13:1077514. doi: 10.3389/fendo.2022.1077514. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36686487 Free PMC article.
-
Transcriptomic Analysis of Ribosome-Bound mRNA in Cortical Neurites In Vivo.J Neurosci. 2017 Sep 6;37(36):8688-8705. doi: 10.1523/JNEUROSCI.3044-16.2017. Epub 2017 Aug 8. J Neurosci. 2017. PMID: 28821669 Free PMC article.
-
Heterogeneity of Hypocretin/Orexin Neurons.Front Neurol Neurosci. 2021;45:61-74. doi: 10.1159/000514964. Epub 2021 May 28. Front Neurol Neurosci. 2021. PMID: 34052814 Free PMC article. Review.
-
Specification of select hypothalamic circuits and innate behaviors by the embryonic patterning gene dbx1.Neuron. 2015 Apr 22;86(2):403-16. doi: 10.1016/j.neuron.2015.03.022. Epub 2015 Apr 9. Neuron. 2015. PMID: 25864637 Free PMC article.
-
Molecular and behavioral profiling of Dbx1-derived neurons in the arcuate, lateral and ventromedial hypothalamic nuclei.Neural Dev. 2016 May 21;11(1):12. doi: 10.1186/s13064-016-0067-9. Neural Dev. 2016. PMID: 27209204 Free PMC article.
References
-
- Bayer L, Eggermann E, Serafin M, Grivel J, Machard D, Muhlethaler M, Jones BE 2005. Opposite effects of noradrenaline and acetylcholine upon hypocretin/orexin versus melanin concentrating hormone neurons in rat hypothalamic slices. Neuroscience 130: 807–811 - PubMed
-
- Bertuzzi S, Porter FD, Pitts A, Kumar M, Agulnick A, Wassif C, Westphal H 1999. Characterization of Lhx9, a novel LIM/homeobox gene expressed by the pioneer neurons in the mouse cerebral cortex. Mech Dev 81: 193–198 - PubMed
-
- Birk OS, Casiano DE, Wassif CA, Cogliati T, Zhao L, Zhao Y, Grinberg A, Huang S, Kreidberg JA, Parker KL, et al. 2000. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 403: 909–913 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
