Leucyl-tRNA synthetase editing domain functions as a molecular rheostat to control codon ambiguity in Mycoplasma pathogens
- PMID: 23431144
- PMCID: PMC3593832
- DOI: 10.1073/pnas.1218374110
Leucyl-tRNA synthetase editing domain functions as a molecular rheostat to control codon ambiguity in Mycoplasma pathogens
Abstract
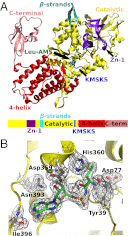
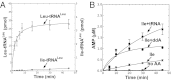
Mycoplasma leucyl-tRNA synthetases (LeuRSs) have been identified in which the connective polypeptide 1 (CP1) amino acid editing domain that clears mischarged tRNAs are missing (Mycoplasma mobile) or highly degenerate (Mycoplasma synoviae). Thus, these enzymes rely on a clearance pathway called pretransfer editing, which hydrolyzes misactivated aminoacyl-adenylate intermediate via a nebulous mechanism that has been controversial for decades. Even as the sole fidelity pathway for clearing amino acid selection errors in the pathogenic M. mobile, pretransfer editing is not robust enough to completely block mischarging of tRNA(Leu), resulting in codon ambiguity and statistical proteins. A high-resolution X-ray crystal structure shows that M. mobile LeuRS structurally overlaps with other LeuRS cores. However, when CP1 domains from different aminoacyl-tRNA synthetases and origins were fused to this common LeuRS core, surprisingly, pretransfer editing was enhanced. It is hypothesized that the CP1 domain evolved as a molecular rheostat to balance multiple functions. These include distal control of specificity and enzyme activity in the ancient canonical core, as well as providing a separate hydrolytic active site for clearing mischarged tRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Molecular modeling study of the editing active site of Escherichia coli leucyl-tRNA synthetase: two amino acid binding sites in the editing domain.Proteins. 2004 Mar 1;54(4):693-704. doi: 10.1002/prot.10300. Proteins. 2004. PMID: 14997565
-
Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity.Proc Natl Acad Sci U S A. 2006 Mar 7;103(10):3586-91. doi: 10.1073/pnas.0507362103. Epub 2006 Feb 27. Proc Natl Acad Sci U S A. 2006. PMID: 16505383 Free PMC article.
-
A paradigm shift for the amino acid editing mechanism of human cytoplasmic leucyl-tRNA synthetase.Biochemistry. 2009 Sep 29;48(38):8958-64. doi: 10.1021/bi901111y. Biochemistry. 2009. PMID: 19702327 Free PMC article.
-
tRNA-independent pretransfer editing by class I leucyl-tRNA synthetase.J Biol Chem. 2009 Feb 6;284(6):3418-24. doi: 10.1074/jbc.M806717200. Epub 2008 Dec 9. J Biol Chem. 2009. PMID: 19068478
-
The balance between pre- and post-transfer editing in tRNA synthetases.FEBS Lett. 2010 Jan 21;584(2):455-9. doi: 10.1016/j.febslet.2009.11.071. FEBS Lett. 2010. PMID: 19941860 Free PMC article. Review.
Cited by
-
tRNA synthetase: tRNA aminoacylation and beyond.Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):461-80. doi: 10.1002/wrna.1224. Epub 2014 Apr 4. Wiley Interdiscip Rev RNA. 2014. PMID: 24706556 Free PMC article. Review.
-
Stress Response and Adaptation Mediated by Amino Acid Misincorporation during Protein Synthesis.Adv Nutr. 2016 Jul 15;7(4):773S-9S. doi: 10.3945/an.115.010991. Print 2016 Jul. Adv Nutr. 2016. PMID: 27422514 Free PMC article. Review.
-
Beware of Mycoplasma Anti-immunoglobulin Strategies.mBio. 2021 Dec 21;12(6):e0197421. doi: 10.1128/mBio.01974-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781733 Free PMC article. Review.
-
Life without tRNAArg-adenosine deaminase TadA: evolutionary consequences of decoding the four CGN codons as arginine in Mycoplasmas and other Mollicutes.Nucleic Acids Res. 2013 Jul;41(13):6531-43. doi: 10.1093/nar/gkt356. Epub 2013 May 8. Nucleic Acids Res. 2013. PMID: 23658230 Free PMC article.
-
Mistranslation can enhance fitness through purging of deleterious mutations.Nat Commun. 2017 May 19;8:15410. doi: 10.1038/ncomms15410. Nat Commun. 2017. PMID: 28524864 Free PMC article.
References
-
- Mascarenhas AP, Martinis SA, An S, Rosen AE, Musier-Forsyth K. 2008. Fidelity mechanisms of the aminoacyl-tRNA synthetases. Protein Engineering, eds RajBhandary UL, Koehrer C (Springer, Berlin), pp 153–200.
-
- Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286(5446):1893–1897. - PubMed
-
- Karkhanis VA, Boniecki MT, Poruri K, Martinis SA. A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J Biol Chem. 2006;281(44):33217–33225. - PubMed
-
- Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

