SOD1 exhibits allosteric frustration to facilitate metal binding affinity
- PMID: 23431152
- PMCID: PMC3593857
- DOI: 10.1073/pnas.1216597110
SOD1 exhibits allosteric frustration to facilitate metal binding affinity
Abstract
Superoxide dismutase-1 (SOD1) is a ubiquitous, Cu and Zn binding, free-radical defense enzyme whose misfolding and aggregation play a potential key role in amyotrophic lateral sclerosis, an invariably fatal neurodegenerative disease. Over 150 mutations in SOD1 have been identified with a familial form of the disease, but it is presently not clear what unifying features, if any, these mutants share to make them pathogenic. Here, we develop several unique computational assays for probing the thermo-mechanical properties of both ALS-associated and rationally designed SOD1 variants. Allosteric interaction-free energies between residues and metals are calculated, and a series of atomic force microscopy experiments are simulated with variable tether positions to quantify mechanical rigidity "fingerprints" for SOD1 variants. Mechanical fingerprinting studies of a series of C-terminally truncated mutants, along with an analysis of equilibrium dynamic fluctuations while varying native constraints, potential energy change upon mutation, frustratometer analysis, and analysis of the coupling between local frustration and metal binding interactions for a glycine scan of 90 residues together, reveal that the apo protein is internally frustrated, that these internal stresses are partially relieved by mutation but at the expense of metal-binding affinity, and that the frustration of a residue is directly related to its role in binding metals. This evidence points to apo SOD1 as a strained intermediate with "self-allostery" for high metal-binding affinity. Thus, the prerequisites for the function of SOD1 as an antioxidant compete with apo state thermo-mechanical stability, increasing the susceptibility of the protein to misfold in the apo state.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


 averaged over residues, as a function truncation length, for the SOD1 variants in A. (A, c) Ribbon schematics of the various truncation mutants, colored blue to red from N to C terminus, labeled by C-terminal residue. (B) Simulated native-basin dynamical fluctuations (RMSF) in explicit simple point charge (SPC) solvent, for Cu,Zn(SS) (black) and E,E(SS) SOD1 monomer (red), along with the experimentally measured ratio of spectral density functions J(ωH)/J(ωN) of obligate monomeric E,E(SS) F50E/G51E/E133Q SOD1 (blue bars) (31). Correlation coefficient is r = 0.78. (C) Simulated RMSF for SOD1 variants E,E G127X (black), E,E(SS) (red), E,E(SH) (blue), and E,E(SH) with the ESL constrained to be natively structured (magenta). The presence of native stress is indicated by the increased disorder of the ZBL upon structuring the ESL (Results). (D) Snapshots of typical structures of E,E(SH) and G127X SOD1 from equilibrium simulations, color coded by the mean RMSF for each residue; RMSF increases from blue to red according to the scale bars shown.
averaged over residues, as a function truncation length, for the SOD1 variants in A. (A, c) Ribbon schematics of the various truncation mutants, colored blue to red from N to C terminus, labeled by C-terminal residue. (B) Simulated native-basin dynamical fluctuations (RMSF) in explicit simple point charge (SPC) solvent, for Cu,Zn(SS) (black) and E,E(SS) SOD1 monomer (red), along with the experimentally measured ratio of spectral density functions J(ωH)/J(ωN) of obligate monomeric E,E(SS) F50E/G51E/E133Q SOD1 (blue bars) (31). Correlation coefficient is r = 0.78. (C) Simulated RMSF for SOD1 variants E,E G127X (black), E,E(SS) (red), E,E(SH) (blue), and E,E(SH) with the ESL constrained to be natively structured (magenta). The presence of native stress is indicated by the increased disorder of the ZBL upon structuring the ESL (Results). (D) Snapshots of typical structures of E,E(SH) and G127X SOD1 from equilibrium simulations, color coded by the mean RMSF for each residue; RMSF increases from blue to red according to the scale bars shown.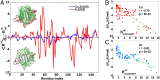
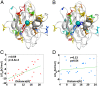
 . A positive number would indicate an increase in frustration upon mutation. Holo state is shown in blue and has an average of +5 contacts; apo state is shown in red and has an average of –22 contacts. (B) Interaction-free energy between a residue’s side chain and the Cu ion, plotted as a function of the E,E(SS) ensemble-averaged number of highly frustrated contacts that residue has (r = –0.79, P = 8e-21). (C) Same as in B but for the Zn ion (r = –0.81, P = 2e-22).
. A positive number would indicate an increase in frustration upon mutation. Holo state is shown in blue and has an average of +5 contacts; apo state is shown in red and has an average of –22 contacts. (B) Interaction-free energy between a residue’s side chain and the Cu ion, plotted as a function of the E,E(SS) ensemble-averaged number of highly frustrated contacts that residue has (r = –0.79, P = 8e-21). (C) Same as in B but for the Zn ion (r = –0.81, P = 2e-22).
References
-
- Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. - PubMed
-
- Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. - PubMed
-
- Weber G. Energetics of ligand binding to proteins. Adv Protein Chem. 1975;29:1–83. - PubMed
-
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57(3):433–443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

