Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy
- PMID: 23431161
- PMCID: PMC3593895
- DOI: 10.1073/pnas.1213676110
Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy
Abstract
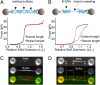
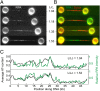
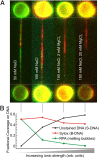
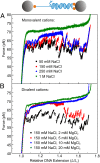
Mechanical stress plays a key role in many genomic processes, such as DNA replication and transcription. The ability to predict the response of double-stranded (ds) DNA to tension is a cornerstone of understanding DNA mechanics. It is widely appreciated that torsionally relaxed dsDNA exhibits a structural transition at forces of ∼65 pN, known as overstretching, whereby the contour length of the molecule increases by ∼70%. Despite extensive investigation, the structural changes occurring in DNA during overstretching are still generating considerable debate. Three mechanisms have been proposed to account for the increase in DNA contour length during overstretching: strand unpeeling, localized base-pair breaking (yielding melting bubbles), and formation of S-DNA (strand unwinding, while base pairing is maintained). Here we show, using a combination of fluorescence microscopy and optical tweezers, that all three structures can exist, uniting the often contradictory dogmas of DNA overstretching. We visualize and distinguish strand unpeeling and melting-bubble formation using an appropriate combination of fluorescently labeled proteins, whereas remaining B-form DNA is accounted for by using specific fluorescent molecular markers. Regions of S-DNA are associated with domains where fluorescent probes do not bind. We demonstrate that the balance between the three structures of overstretched DNA is governed by both DNA topology and local DNA stability. These findings enhance our knowledge of DNA mechanics and stability, which are of fundamental importance to understanding how proteins modify the physical state of DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):3865-70. doi: 10.1073/pnas.1213740110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431154 Free PMC article.
-
Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18231-6. doi: 10.1073/pnas.0904322106. Epub 2009 Oct 19. Proc Natl Acad Sci U S A. 2009. PMID: 19841258 Free PMC article.
-
Force-induced melting and S-DNA pathways for DNA overstretching exhibit distinct kinetics.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1183. doi: 10.1093/nar/gkae1183. Nucleic Acids Res. 2025. PMID: 39657777 Free PMC article.
-
Thermodynamics of DNA interactions from single molecule stretching experiments.Acc Chem Res. 2002 Mar;35(3):159-66. doi: 10.1021/ar010045k. Acc Chem Res. 2002. PMID: 11900519 Review.
-
Denaturation transition of stretched DNA.Biochem Soc Trans. 2013 Apr;41(2):639-45. doi: 10.1042/BST20120298. Biochem Soc Trans. 2013. PMID: 23514169 Review.
Cited by
-
Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase.Cell. 2019 Jul 25;178(3):600-611.e16. doi: 10.1016/j.cell.2019.06.032. Cell. 2019. PMID: 31348887 Free PMC article.
-
Differential contribution of basic residues to HIV-1 nucleocapsid protein's nucleic acid chaperone function and retroviral replication.Nucleic Acids Res. 2014 Feb;42(4):2525-37. doi: 10.1093/nar/gkt1227. Epub 2013 Nov 28. Nucleic Acids Res. 2014. PMID: 24293648 Free PMC article.
-
Dynamic topology of double-stranded telomeric DNA studied by single-molecule manipulation in vitro.Nucleic Acids Res. 2020 Jul 9;48(12):6458-6470. doi: 10.1093/nar/gkaa479. Nucleic Acids Res. 2020. PMID: 32496520 Free PMC article.
-
Multi-plateau force-extension curves of long double-stranded DNA molecules.bioRxiv [Preprint]. 2025 Feb 28:2023.03.12.532320. doi: 10.1101/2023.03.12.532320. bioRxiv. 2025. Update in: ACS Omega. 2025 Apr 10;10(15):14816-14825. doi: 10.1021/acsomega.4c09605. PMID: 40060417 Free PMC article. Updated. Preprint.
-
Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy.Nucleic Acids Res. 2016 May 19;44(9):3971-88. doi: 10.1093/nar/gkw237. Epub 2016 Apr 16. Nucleic Acids Res. 2016. PMID: 27085806 Free PMC article.
References
-
- Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444(7117):387–390. - PubMed
-
- Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271(5250):795–799. - PubMed
-
- Cluzel P, et al. DNA: An extensible molecule. Science. 1996;271(5250):792–794. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

