Comparative transcriptomics of early dipteran development
- PMID: 23432914
- PMCID: PMC3616871
- DOI: 10.1186/1471-2164-14-123
Comparative transcriptomics of early dipteran development
Abstract
Background: Modern sequencing technologies have massively increased the amount of data available for comparative genomics. Whole-transcriptome shotgun sequencing (RNA-seq) provides a powerful basis for comparative studies. In particular, this approach holds great promise for emerging model species in fields such as evolutionary developmental biology (evo-devo).
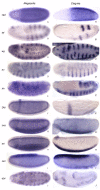
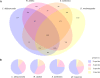
Results: We have sequenced early embryonic transcriptomes of two non-drosophilid dipteran species: the moth midge Clogmia albipunctata, and the scuttle fly Megaselia abdita. Our analysis includes a third, published, transcriptome for the hoverfly Episyrphus balteatus. These emerging models for comparative developmental studies close an important phylogenetic gap between Drosophila melanogaster and other insect model systems. In this paper, we provide a comparative analysis of early embryonic transcriptomes across species, and use our data for a phylogenomic re-evaluation of dipteran phylogenetic relationships.
Conclusions: We show how comparative transcriptomics can be used to create useful resources for evo-devo, and to investigate phylogenetic relationships. Our results demonstrate that de novo assembly of short (Illumina) reads yields high-quality, high-coverage transcriptomic data sets. We use these data to investigate deep dipteran phylogenetic relationships. Our results, based on a concatenation of 160 orthologous genes, provide support for the traditional view of Clogmia being the sister group of Brachycera (Megaselia, Episyrphus, Drosophila), rather than that of Culicomorpha (which includes mosquitoes and blackflies).
Figures



Similar articles
-
Maternal co-ordinate gene regulation and axis polarity in the scuttle fly Megaselia abdita.PLoS Genet. 2015 Mar 10;11(3):e1005042. doi: 10.1371/journal.pgen.1005042. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25757102 Free PMC article.
-
SuperFly: a comparative database for quantified spatio-temporal gene expression patterns in early dipteran embryos.Nucleic Acids Res. 2015 Jan;43(Database issue):D751-5. doi: 10.1093/nar/gku1142. Epub 2014 Nov 17. Nucleic Acids Res. 2015. PMID: 25404137 Free PMC article.
-
Quantitative system drift compensates for altered maternal inputs to the gap gene network of the scuttle fly Megaselia abdita.Elife. 2015 Jan 5;4:e04785. doi: 10.7554/eLife.04785. Elife. 2015. PMID: 25560971 Free PMC article.
-
Comparative Transcriptomes and EVO-DEVO Studies Depending on Next Generation Sequencing.Comput Math Methods Med. 2015;2015:896176. doi: 10.1155/2015/896176. Epub 2015 Oct 12. Comput Math Methods Med. 2015. PMID: 26543497 Free PMC article. Review.
-
[Progress on genome sequencing of Dipteran insects].Yi Chuan. 2020 Nov 20;42(11):1093-1109. doi: 10.16288/j.yczz.20-130. Yi Chuan. 2020. PMID: 33229316 Review. Chinese.
Cited by
-
Maternal co-ordinate gene regulation and axis polarity in the scuttle fly Megaselia abdita.PLoS Genet. 2015 Mar 10;11(3):e1005042. doi: 10.1371/journal.pgen.1005042. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25757102 Free PMC article.
-
Phylogenomics Identifies an Ancestral Burst of Gene Duplications Predating the Diversification of Aphidomorpha.Mol Biol Evol. 2020 Mar 1;37(3):730-756. doi: 10.1093/molbev/msz261. Mol Biol Evol. 2020. PMID: 31702774 Free PMC article.
-
Transcriptome Analysis of the Oriental Fruit Fly Bactrocera dorsalis Early Embryos.Insects. 2020 May 23;11(5):323. doi: 10.3390/insects11050323. Insects. 2020. PMID: 32456171 Free PMC article.
-
The dorsoventral patterning of Musca domestica embryos: insights into BMP/Dpp evolution from the base of the lower cyclorraphan flies.Evodevo. 2018 May 16;9:13. doi: 10.1186/s13227-018-0102-5. eCollection 2018. Evodevo. 2018. PMID: 29796243 Free PMC article.
-
Genomic Resources for the Scuttle Fly Megaselia abdita: A Model Organism for Comparative Developmental Studies in Flies.bioRxiv [Preprint]. 2025 Feb 4:2025.01.13.631075. doi: 10.1101/2025.01.13.631075. bioRxiv. 2025. PMID: 39868096 Free PMC article. Preprint.
References
-
- Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, Raff EC, Pace NR, Raff RA. Molecular Phylogeny of the Animal Kingdom. Science. 1988;239:748–53. - PubMed
-
- Aguinaldo AMA, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–93. - PubMed
-
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–75. - PubMed
-
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

