Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation
- PMID: 23434372
- PMCID: PMC3628298
- DOI: 10.1016/j.molcel.2013.01.022
Class IA PI3K p110β subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation
Abstract
Autophagy is an evolutionarily conserved membrane trafficking process. Induction of autophagy in response to nutrient limitation or cellular stress occurs by similar mechanisms in organisms from yeast to mammals. Unlike yeast, metazoan cells rely more on growth factor signaling for a wide variety of cellular activities including nutrient uptake. How growth factor availability regulates autophagy is poorly understood. Here we show that, upon growth factor limitation, the p110β catalytic subunit of the class IA phosphoinositide 3-kinases (PI3Ks) dissociates from growth factor receptor complexes and increases its interaction with the small GTPase Rab5. This p110β-Rab5 association maintains Rab5 in its guanosine triphosphate (GTP)-bound state and enhances the Rab5-Vps34 interaction that promotes autophagy. p110β mutants that fail to interact with Rab5 are defective in autophagy promotion. Hence, in mammalian cells, p110β acts as a molecular sensor for growth factor availability and induces autophagy by activating a Rab5-mediated signaling cascade.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


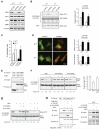
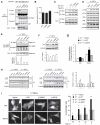
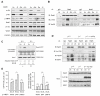

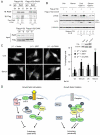
References
-
- Barbieri MA, Li G, Colombo MI, Stahl PD. Rab5, an early acting endosomal GTPase, supports in vitro endosome fusion without GTP hydrolysis. J Biol Chem. 1994;269:18720–18722. - PubMed
-
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. - PubMed
-
- Carpenter CL, Duckworth BC, Auger KR, Cohen B, Schaffhausen BS, Cantley LC. Purification and characterization of phosphoinositide 3-kinase from rat liver. J Biol Chem. 1990;265:19704–19711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA136754/CA/NCI NIH HHS/United States
- R01 CA136754/CA/NCI NIH HHS/United States
- GM97355/GM/NIGMS NIH HHS/United States
- GM74692/GM/NIGMS NIH HHS/United States
- AG039632/AG/NIA NIH HHS/United States
- GM55692/GM/NIGMS NIH HHS/United States
- R01 DK062722/DK/NIDDK NIH HHS/United States
- R01 AG039632/AG/NIA NIH HHS/United States
- CA129536/CA/NCI NIH HHS/United States
- R01 GM055692/GM/NIGMS NIH HHS/United States
- R01 GM097355/GM/NIGMS NIH HHS/United States
- DK62722/DK/NIDDK NIH HHS/United States
- R01 CA129536/CA/NCI NIH HHS/United States
- R01 GM074692/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

