Allosteric inhibition of hypoxia inducible factor-2 with small molecules
- PMID: 23434853
- PMCID: PMC3604136
- DOI: 10.1038/nchembio.1185
Allosteric inhibition of hypoxia inducible factor-2 with small molecules
Abstract
Hypoxia inducible factors (HIFs) are heterodimeric transcription factors induced in many cancers where they frequently promote the expression of protumorigenic pathways. Though transcription factors are typically considered 'undruggable', the PAS-B domain of the HIF-2α subunit contains a large cavity within its hydrophobic core that offers a unique foothold for small-molecule regulation. Here we identify artificial ligands that bind within this pocket and characterize the resulting structural and functional changes caused by binding. Notably, these ligands antagonize HIF-2 heterodimerization and DNA-binding activity in vitro and in cultured cells, reducing HIF-2 target gene expression. Despite the high sequence identity between HIF-2α and HIF-1α, these ligands are highly selective and do not affect HIF-1 function. These chemical tools establish the molecular basis for selective regulation of HIF-2, providing potential therapeutic opportunities to intervene in HIF-2-driven tumors, such as renal cell carcinomas.
Conflict of interest statement
R.K.B., K.H.G., T.H.S., J.K. and U.K.T. have received stock options and other financial compensation from Peloton Therapeutics Inc.
Figures

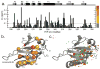

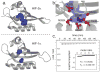

References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

