Role of PelF in pel polysaccharide biosynthesis in Pseudomonas aeruginosa
- PMID: 23435893
- PMCID: PMC3623137
- DOI: 10.1128/AEM.03666-12
Role of PelF in pel polysaccharide biosynthesis in Pseudomonas aeruginosa
Abstract
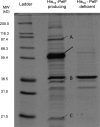
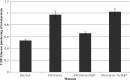
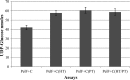
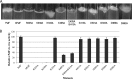
Pseudomonas aeruginosa produces three exopolysaccharides, Psl, Pel, and alginate, that play vital roles in biofilm formation. Pel is a glucose-rich, cellulose-like exopolysaccharide. The essential Pel biosynthesis proteins are encoded by seven genes, pelA to pelG. Bioinformatics analysis suggests that PelF is a cytosolic glycosyltransferase. Here, experimental evidence was provided to support this PelF function. A UDP-glucose dehydrogenase-based assay was developed to quantify UDP-glucose. UDP-glucose was proposed as the substrate for PelF. The isogenic pelF deletion mutant accumulated 1.8 times more UDP-glucose in its cytosol than the wild type. This suggested that PelF, which was found localized in the cystosol, uses UDP-glucose as substrate. Additionally, in vitro experiments confirmed that PelF uses UDP-glucose as substrate. To analyze the functional roles of conserved residues in PelF, site-directed mutagenesis was performed. The presence of the EX7E motif is characteristic for various glycosyltransferase families, and in PelF, E405/E413 are the conserved residues in this motif. Replacement of E405 with A resulted in a reduction of PelF activity to 30.35% ± 3.15% (mean ± standard deviation) of the wild-type level, whereas replacement of the second E, E413, with A did not produce a significant change in the activity of PelF. Moreover, replacement of both E residues did not result in a loss of PelF function, but replacement of the conserved R325 or K330 with A resulted in a complete loss of PelF activity. Overall, our data show that PelF is a soluble glycosyltransferase that uses UDP-glucose as the substrate for Pel synthesis and that conserved residues R325 and K330 are important for the activity of PelF.
Figures


References
-
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407: 762–764 - PubMed
-
- Drenkard E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5: 1213–1219 - PubMed
-
- Chitnis CE, Ohman DE. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8: 583–593 - PubMed
-
- Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51: 675–690 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

