Theory of the Protein Equilibrium Population Snapshot by H/D Exchange Electrospray Ionization Mass Spectrometry (PEPS-HDX-ESI-MS) Method used to obtain Protein Folding Energies/Rates and Selected Supporting Experimental Evidence
- PMID: 23436981
- PMCID: PMC3578662
- DOI: 10.1016/j.ijms.2012.10.010
Theory of the Protein Equilibrium Population Snapshot by H/D Exchange Electrospray Ionization Mass Spectrometry (PEPS-HDX-ESI-MS) Method used to obtain Protein Folding Energies/Rates and Selected Supporting Experimental Evidence
Abstract
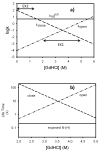
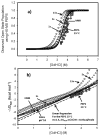
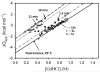
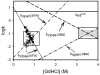
Protein equilibrium snapshot by hydrogen/deuterium exchange electrospray ionization mass spectrometry (PEPS-HDX-ESI-MS or PEPS) is a method recently introduced for estimating protein folding energies and rates. Herein we describe the basis for this method using both theory and new experiments. Benchmark experiments were conducted using ubiquitin because of the availability of reference data for folding and unfolding rates from NMR studies. A second set of experiments was also conducted to illustrate the surprising resilience of the PEPS to changes in HDX time, using staphylococcal nuclease and time frames ranging from a few seconds to several minutes. Theory suggests that PEPS experiments should be conducted at relatively high denaturant concentrations, where the protein folding/unfolding rates are slow with respect to HDX and the life times of both the closed and open states are long enough to be sampled experimentally. Upon deliberate denaturation, changes in folding/unfolding are correlated with associated changes in the ESI-MS signal upon fast HDX. When experiments are done quickly, typically within a few seconds, ESI-MS signals, corresponding to the equilibrium population of the native (closed) and denatured (open) states can both be detected. The interior of folded proteins remains largely un-exchanged. Amongst MS methods, the simultaneous detection of both states in the spectrum is unique to PEPS and provides a "snapshot" of these populations. The associated ion intensities are used to estimate the protein folding equilibrium constant (or the free energy change, ΔG). Linear extrapolation method (LEM) plots of derived ΔG values for each denaturant concentration can then be used to calculate ΔG in the absence of denaturant, ΔG(H(2)O). In accordance with the requirement for detection of signals for both the folded and unfolded states, this theoretical framework predicts that PEPS experiments work best at the middle of the denaturation curve where natured and denatured protein molecules are equilibrated at easily detectable ratios, namely 1:1. It also requires that closed and open states have lifetimes measurable in the time frame of the HDX experiment. Because both conditions are met by PEPS, these measurements can provide an accurate assessment of closed/open state populations and thus protein folding energies/rates.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
