Expression and purification of functional human mu opioid receptor from E.coli
- PMID: 23437147
- PMCID: PMC3578875
- DOI: 10.1371/journal.pone.0056500
Expression and purification of functional human mu opioid receptor from E.coli
Abstract
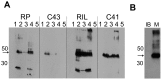
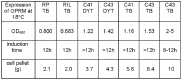
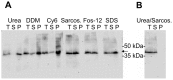
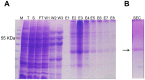
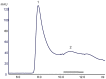
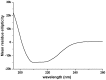
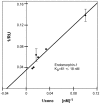
N-terminally his-tagged human mu opioid receptor, a G protein-coupled receptor was produced in E.coli employing synthetic codon-usage optimized constructs. The receptor was expressed in inclusion bodies and membrane-inserted in different E.coli strains. By optimizing the expression conditions the expression level for the membrane-integrated receptor was raised to 0.3-0.5 mg per liter of culture. Milligram quantities of receptor could be enriched by affinity chromatography from IPTG induced cultures grown at 18°C. By size exclusion chromatography the protein fraction with the fraction of alpha-helical secondary structure expected for a 7-TM receptor was isolated, by CD-spectroscopy an alpha-helical content of ca. 45% was found for protein solubilised in the detergent Fos-12. Receptor in Fos-12 micelles was shown to bind endomorphin-1 with a K(D) of 61 nM. A final yield of 0.17 mg functional protein per liter of culture was obtained.
Conflict of interest statement
Figures

References
-
- Flower DR (1999) Modelling G-protein-coupled receptors for drug design. Biochim Biophys Acta 1422: 207–234. - PubMed
-
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63: 1256–1272. - PubMed
-
- Schlyer S, Horuk R (2006) I want a new drug: G-protein-coupled receptors in drug development. Drug Discov Today 11: 481–493. - PubMed
-
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S (2002) Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett 520: 97–101. - PubMed
-
- Tyndall JD, Sandilya R (2005) GPCR agonists and antagonists in the clinic. Med Chem 1: 405–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

