"Function-first" lead discovery: mode of action profiling of natural product libraries using image-based screening
- PMID: 23438757
- PMCID: PMC3584419
- DOI: 10.1016/j.chembiol.2012.12.007
"Function-first" lead discovery: mode of action profiling of natural product libraries using image-based screening
Abstract
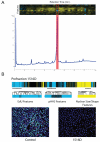
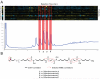
Cytological profiling is a high-content image-based screening technology that provides insight into the mode of action (MOA) for test compounds by directly measuring hundreds of phenotypic cellular features. We have extended this recently reported technology to the mechanistic characterization of unknown natural products libraries for the direct prediction of compound MOAs at the primary screening stage. By analyzing a training set of commercial compounds of known mechanism and comparing these profiles to those obtained from natural product library members, we have successfully annotated extracts based on MOA, dereplicated known compounds based on biological similarity to the training set, and identified and predicted the MOA of a unique family of iron siderophores. Coupled with traditional analytical techniques, cytological profiling provides an avenue for the creation of "function-first" approaches to natural products discovery.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

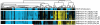

References
-
- Ando T, Hirayama K, Takahashi R, Horino I, Etoh Y, Morioka H, Shibai H, Murai A. Cosmomycin D, a New Anthracycline Antibiotic. Agr. Biol. Chem. 1985;49:259–262.
-
- Arora SK. Molecular structure of heliomycin, an inhibitor of RNA synthesis. J. Antibiot. 1985;38:113–115. - PubMed
-
- Brockmann H, Schmidt-Kastner G. Valinomycin I, XXVII. Mitteil. über Antibiotica aus Actinomyceten. Chem. Ber. 1955;88:57–61.
-
- Cipollone a, Berettoni M, Bigioni M, Binaschi M, Cermele C, Monteagudo E, Olivieri L, Palomba D, Animati F, Goso C, et al. Novel anthracycline oligosaccharides: influence of chemical modifications of the carbohydrate moiety on biological activity. Bioorgan. Med. Chem. 2002;10:1459–1470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

