CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes
- PMID: 23439366
- PMCID: PMC3737325
- DOI: 10.4161/rna.24022
CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes
Abstract
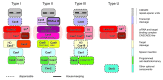
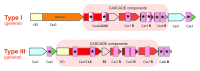
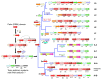
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR-associated genes) is an adaptive immunity system in bacteria and archaea that functions via a distinct self-non-self recognition mechanism that is partially analogous to the mechanism of eukaryotic RNA interference (RNAi). The CRISPR-Cas system incorporates fragments of virus or plasmid DNA into the CRISPR repeat cassettes and employs the processed transcripts of these spacers as guide RNAs to cleave the cognate foreign DNA or RNA. The Cas proteins, however, are not homologous to the proteins involved in RNAi and comprise numerous, highly diverged families. The majority of the Cas proteins contain diverse variants of the RNA recognition motif (RRM), a widespread RNA-binding domain. Despite the fast evolution that is typical of the cas genes, the presence of diverse versions of the RRM in most Cas proteins provides for a simple scenario for the evolution of the three distinct types of CRISPR-cas systems. In addition to several proteins that are directly implicated in the immune response, the cas genes encode a variety of proteins that are homologous to prokaryotic toxins that typically possess nuclease activity. The predicted toxins associated with CRISPR-Cas systems include the essential Cas2 protein, proteins of COG1517 that, in addition to a ligand-binding domain and a helix-turn-helix domain, typically contain different nuclease domains and several other predicted nucleases. The tight association of the CRISPR-Cas immunity systems with predicted toxins that, upon activation, would induce dormancy or cell death suggests that adaptive immunity and dormancy/suicide response are functionally coupled. Such coupling could manifest in the persistence state being induced and potentially providing conditions for more effective action of the immune system or in cell death being triggered when immunity fails.
Keywords: CRISPR-Cas; RRM domain; adaptive immunity; dormancy; innate immunity; programmed cell death.
Figures

References
-
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
