Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection
- PMID: 23439496
- PMCID: PMC3666629
- DOI: 10.1038/mt.2013.31
Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection
Abstract
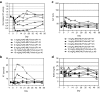
RNA interference (RNAi)-based therapeutics have the potential to treat chronic hepatitis B virus (HBV) infection in a fundamentally different manner than current therapies. Using RNAi, it is possible to knock down expression of viral RNAs including the pregenomic RNA from which the replicative intermediates are derived, thus reducing viral load, and the viral proteins that result in disease and impact the immune system's ability to eliminate the virus. We previously described the use of polymer-based Dynamic PolyConjugate (DPC) for the targeted delivery of siRNAs to hepatocytes. Here, we first show in proof-of-concept studies that simple coinjection of a hepatocyte-targeted, N-acetylgalactosamine-conjugated melittin-like peptide (NAG-MLP) with a liver-tropic cholesterol-conjugated siRNA (chol-siRNA) targeting coagulation factor VII (F7) results in efficient F7 knockdown in mice and nonhuman primates without changes in clinical chemistry or induction of cytokines. Using transient and transgenic mouse models of HBV infection, we show that a single coinjection of NAG-MLP with potent chol-siRNAs targeting conserved HBV sequences resulted in multilog repression of viral RNA, proteins, and viral DNA with long duration of effect. These results suggest that coinjection of NAG-MLP and chol-siHBVs holds great promise as a new therapeutic for patients chronically infected with HBV.
Figures



References
-
- Seeger C, Zoulim F., and, Mason WS.2007Hepadnaviruses Fields BN, Knipe DM., and, Howley PM.eds.). Fields Virology5th edn. Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia
-
- Chen JD, Yang HI, Iloeje UH, You SL, Lu SN, Wang LY, Risk Evaluation of Viral Load Elevation and Associated Liver Disease/Cancer in HBV (REVEAL-HBV) Study Group et al. Carriers of inactive hepatitis B virus are still at risk for hepatocellular carcinoma and liver-related death. Gastroenterology. 2010;138:1747–1754. - PubMed
-
- Yuen MF., and, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. J Gastroenterol Hepatol. 2011;26 Suppl 1:138–143. - PubMed
-
- Funk ML, Rosenberg DM., and, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepat. 2002;9:52–61. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

