Biochemical and biophysical characterization of the sialyl-/hexosyltransferase synthesizing the meningococcal serogroup W135 heteropolysaccharide capsule
- PMID: 23439648
- PMCID: PMC3636861
- DOI: 10.1074/jbc.M113.452276
Biochemical and biophysical characterization of the sialyl-/hexosyltransferase synthesizing the meningococcal serogroup W135 heteropolysaccharide capsule
Abstract
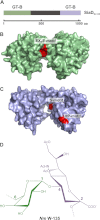
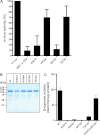
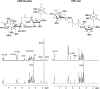
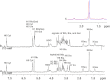
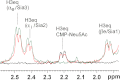
Neisseria meningitidis (Nm) is a leading cause of bacterial meningitis and sepsis. Crucial virulence determinants of pathogenic Nm strains are the polysaccharide capsules that support invasion by hindering complement attack. In NmW-135 and NmY the capsules are built from the repeating units (→ 6)-α-D-Gal-(1 → 4)-α-Neu5Ac-(2 →)n and (→ 6)-α-D-Glc-(1 → 4)-α-Neu5Ac-(2 →)n, respectively. These unusual heteropolymers represent unique examples of a conjugation between sialic acid and hexosyl-sugars in a polymer chain. Moreover, despite the various catalytic strategies needed for sialic acid and hexose transfer, single enzymes (SiaDW-135/Y) have been identified to form these heteropolymers. Here we used SiaDW-135 as a model system to delineate structure-function relationships. In size exclusion chromatography active SiaDW-135 migrated as a monomer. Fold recognition programs suggested two separate glycosyltransferase domains, both containing a GT-B-fold. Based on conserved motifs predicted folds could be classified as a hexosyl- and sialyltransferase. To analyze enzyme properties and interplay of the two identified glycosyltransferase domains, saturation transfer difference NMR and mutational studies were carried out. Simultaneous and independent binding of UDP-Gal and CMP-Sia was seen in the absence of an acceptor as well as when the catalytic cycle was allowed to proceed. Enzyme variants with only one functionality were generated by site-directed mutagenesis and shown to complement each other in trans when combined in an in vitro test system. Together the data strongly suggests that SiaDW-135 has evolved by fusion of two independent ancestral genes encoding sialyl- and galactosyltransferase activity.
Figures





References
-
- Rosenstein N. E., Perkins B. A., Stephens D. S., Popovic T., Hughes J. M. (2001) Meningococcal disease. N. Engl. J. Med. 344, 1378–1388 - PubMed
-
- Stephens D. S., Greenwood B., Brandtzaeg P. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 - PubMed
-
- Raymond N. J., Reeves M., Ajello G., Baughman W., Gheesling L. L., Carlone G. M., Wenger J. D., Stephens D. S. (1997) Molecular epidemiology of sporadic (endemic) serogroup C meningococcal disease. J. Infect. Dis. 176, 1277–1284 - PubMed
-
- Taha M. K., Achtman M., Alonso J. M., Greenwood B., Ramsay M., Fox A., Gray S., Kaczmarski E. (2000) Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356, 2159. - PubMed
-
- Decosas J., Koama J. B. (2002) Chronicle of an outbreak foretold. Meningococcal meningitis W135 in Burkina Faso. Lancet Infect. Dis. 2, 763–765 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

