Detection of replication-defective hepatitis A virus based on the correlation between real-time polymerase chain reaction and ELISA in situ results
- PMID: 23440112
- PMCID: PMC3974324
- DOI: 10.1590/s0074-02762013000100006
Detection of replication-defective hepatitis A virus based on the correlation between real-time polymerase chain reaction and ELISA in situ results
Abstract
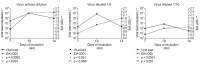
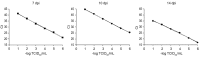
ELISA in situ can be used to titrate hepatitis A virus (HAV) particles and real-time polymerase chain reaction (RT-PCR) has been shown to be a fast method to quantify the HAV genome. Precise quantification of viral concentration is necessary to distinguish between infectious and non-infectious particles. The purpose of this study was to compare cell culture and RT-PCR quantification results and determine whether HAV genome quantification can be correlated with infectivity. For this purpose, three stocks of undiluted, five-fold diluted and 10-fold diluted HAV were prepared to inoculate cells in a 96-well plate. Monolayers were then incubated for seven, 10 and 14 days and the correlation between the ELISA in situ and RT-PCR results was evaluated. At 10 days post-incubation, the highest viral load was observed in all stocks of HAV via RT-PCR (10(5) copies/mL) (p = 0.0002), while ELISA revealed the highest quantity of particles after 14 days (optical density = 0.24, p < 0.001). At seven days post-infection, there was a significant statistical correlation between the results of the two methods, indicating equivalents titres of particles and HAV genome during this period of infection. The results reported here indicate that the duration of growth of HAV in cell culture must be taken into account to correlate genome quantification with infectivity.
Figures
Similar articles
-
Development of a Real-Time Cell Analysis (RTCA) Method as a Fast and Accurate Method for Detecting Infectious Particles of the Adapted Strain of Hepatitis A Virus.Front Cell Infect Microbiol. 2018 Sep 25;8:335. doi: 10.3389/fcimb.2018.00335. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30319992 Free PMC article.
-
Development of duplex RT-PCR-ELISA for the simultaneous detection of hepatitis A virus and hepatitis E virus.J Virol Methods. 2011 Jul;175(1):137-40. doi: 10.1016/j.jviromet.2011.04.030. Epub 2011 May 4. J Virol Methods. 2011. PMID: 21565222
-
Application of viability PCR to discriminate the infectivity of hepatitis A virus in food samples.Int J Food Microbiol. 2015 May 18;201:1-6. doi: 10.1016/j.ijfoodmicro.2015.02.012. Epub 2015 Feb 16. Int J Food Microbiol. 2015. PMID: 25720326
-
Discrimination of infectious hepatitis A viruses by propidium monoazide real-time RT-PCR.Food Environ Virol. 2012 Mar;4(1):21-5. doi: 10.1007/s12560-011-9074-5. Epub 2011 Dec 17. Food Environ Virol. 2012. PMID: 23412764
-
Comparative evaluation of a novel TaqMan real-time reverse transcription-polymerase chain reaction assay for hepatitis A virus detection.J Int Med Res. 2013 Apr;41(2):427-34. doi: 10.1177/0300060513476434. Epub 2013 Mar 7. J Int Med Res. 2013. PMID: 23569019
Cited by
-
Inter- and Intra-Host Nucleotide Variations in Hepatitis A Virus in Culture and Clinical Samples Detected by Next-Generation Sequencing.Viruses. 2018 Nov 9;10(11):619. doi: 10.3390/v10110619. Viruses. 2018. PMID: 30423964 Free PMC article.
References
-
- de Paula VS, Diniz-Mendes L, Villar LM, Luz SL, Silva LA, Jesus MS, da Silva NM, Gaspar AM. Hepatitis A virus in environmental water samples from the Amazon Basin. Water Res. 2007;41:1169–1176. - PubMed
-
- de Paula VS, Perse AS, Amado LA, de Morais LM, de Lima SM, Tourinho RS, Gaspar AM, Pinto MA. Kinetics of hepatitis A virus replication in vivo and in vitro using negative-strand quantitative PCR. Eur J Clin Microbiol Infect Dis. 2009;28:1167–1176. - PubMed
-
- Donia D, Bonanni E, Diaco L, Divizia M. Statistical correlation between enterovirus genome copy numbers and infectious viral particles in wastewater samples. Lett Appl Microbiol. 2009;50:237–240. - PubMed
-
- Dudek T, Knipe DM. Replication-defective viruses as vaccines and vaccine vectors. Virology. 2006;344:230–239. - PubMed
-
- Forcic D, Kosutic-Gulija T, Santak M, Jug R, Ivancic-Jelecki J, Markusic M, Mazuran R. Comparisons of mumps virus potency estimates obtained by 50% cell culture infective dose assay and plaque assay. Vaccine. 2009;28:1887–1892. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

