The virtual brain integrates computational modeling and multimodal neuroimaging
- PMID: 23442172
- PMCID: PMC3696923
- DOI: 10.1089/brain.2012.0120
The virtual brain integrates computational modeling and multimodal neuroimaging
Abstract
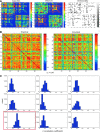
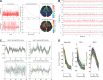
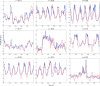
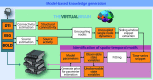
Brain function is thought to emerge from the interactions among neuronal populations. Apart from traditional efforts to reproduce brain dynamics from the micro- to macroscopic scales, complementary approaches develop phenomenological models of lower complexity. Such macroscopic models typically generate only a few selected-ideally functionally relevant-aspects of the brain dynamics. Importantly, they often allow an understanding of the underlying mechanisms beyond computational reproduction. Adding detail to these models will widen their ability to reproduce a broader range of dynamic features of the brain. For instance, such models allow for the exploration of consequences of focal and distributed pathological changes in the system, enabling us to identify and develop approaches to counteract those unfavorable processes. Toward this end, The Virtual Brain (TVB) ( www.thevirtualbrain.org ), a neuroinformatics platform with a brain simulator that incorporates a range of neuronal models and dynamics at its core, has been developed. This integrated framework allows the model-based simulation, analysis, and inference of neurophysiological mechanisms over several brain scales that underlie the generation of macroscopic neuroimaging signals. In this article, we describe how TVB works, and we present the first proof of concept.
Figures


References
-
- Agrafiotis DK. Stochastic proximity embedding. J Comput Chem. 2003;24:1215–1221. - PubMed
-
- Amari S.-i. “Dynamics of pattern formation in lateral-inhibition type neural fields.”. Biological cybernetics. 1977;27:77–87. - PubMed
-
- Assisi CG. Jirsa VK, et al. Synchrony and clustering in heterogeneous networks with global coupling and parameter dispersion. Phys Rev Lett. 2005;94:018106. - PubMed
-
- Attwell D. Iadecola C. The neural basis of functional brain imaging signals. Trends Neurosci. 2002;25:621–625. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
