Transposon activation mutagenesis as a screening tool for identifying resistance to cancer therapeutics
- PMID: 23442791
- PMCID: PMC3598783
- DOI: 10.1186/1471-2407-13-93
Transposon activation mutagenesis as a screening tool for identifying resistance to cancer therapeutics
Abstract
Background: The development of resistance to chemotherapies represents a significant barrier to successful cancer treatment. Resistance mechanisms are complex, can involve diverse and often unexpected cellular processes, and can vary with both the underlying genetic lesion and the origin or type of tumor. For these reasons developing experimental strategies that could be used to understand, identify and predict mechanisms of resistance in different malignant cells would be a major advance.
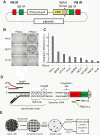
Methods: Here we describe a gain-of-function forward genetic approach for identifying mechanisms of resistance. This approach uses a modified piggyBac transposon to generate libraries of mutagenized cells, each containing transposon insertions that randomly activate nearby gene expression. Genes of interest are identified using next-gen high-throughput sequencing and barcode multiplexing is used to reduce experimental cost.
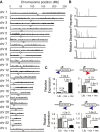
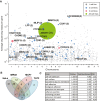
Results: Using this approach we successfully identify genes involved in paclitaxel resistance in a variety of cancer cell lines, including the multidrug transporter ABCB1, a previously identified major paclitaxel resistance gene. Analysis of co-occurring transposons integration sites in single cell clone allows for the identification of genes that might act cooperatively to produce drug resistance a level of information not accessible using RNAi or ORF expression screening approaches.
Conclusion: We have developed a powerful pipeline to systematically discover drug resistance in mammalian cells in vitro. This cost-effective approach can be readily applied to different cell lines, to identify canonical or context specific resistance mechanisms. Its ability to probe complex genetic context and non-coding genomic elements as well as cooperative resistance events makes it a good complement to RNAi or ORF expression based screens.
Figures


References
-
- Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9(6):389–399. - PubMed
-
- Dorssers LC, van Agthoven T, Dekker A, van Agthoven TL, Kok EM. Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: identification of bcar-1, a common integration site. Mol Endocrinol. 1993;7(7):870–878. doi: 10.1210/me.7.7.870. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

