Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts
- PMID: 23442824
- PMCID: PMC3635915
- DOI: 10.1186/1741-7007-11-15
Quantifying the contribution of chromatin dynamics to stochastic gene expression reveals long, locus-dependent periods between transcriptional bursts
Abstract
Background: A number of studies have established that stochasticity in gene expression may play an important role in many biological phenomena. This therefore calls for further investigations to identify the molecular mechanisms at stake, in order to understand and manipulate cell-to-cell variability. In this work, we explored the role played by chromatin dynamics in the regulation of stochastic gene expression in higher eukaryotic cells.
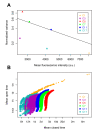
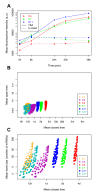
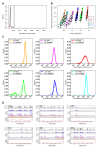
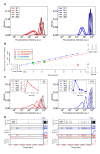
Results: For this purpose, we generated isogenic chicken-cell populations expressing a fluorescent reporter integrated in one copy per clone. Although the clones differed only in the genetic locus at which the reporter was inserted, they showed markedly different fluorescence distributions, revealing different levels of stochastic gene expression. Use of chromatin-modifying agents showed that direct manipulation of chromatin dynamics had a marked effect on the extent of stochastic gene expression. To better understand the molecular mechanism involved in these phenomena, we fitted these data to a two-state model describing the opening/closing process of the chromatin. We found that the differences between clones seemed to be due mainly to the duration of the closed state, and that the agents we used mainly seem to act on the opening probability.
Conclusions: In this study, we report biological experiments combined with computational modeling, highlighting the importance of chromatin dynamics in stochastic gene expression. This work sheds a new light on the mechanisms of gene expression in higher eukaryotic cells, and argues in favor of relatively slow dynamics with long (hours to days) periods of quiet state.
Figures


References
-
- Kupiec JJ. A probabilistic theory for cell differentiation, embryonic mortality and DNA C-value paradox. Speculations in Science and Technology. 1983;6:471–478.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

