Absolute hydration free energies of blocked amino acids: implications for protein solvation and stability
- PMID: 23442867
- PMCID: PMC3552266
- DOI: 10.1016/j.bpj.2012.12.008
Absolute hydration free energies of blocked amino acids: implications for protein solvation and stability
Abstract
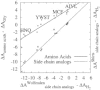
Most proteins perform their function in aqueous solution. The interactions with water determine the stability of proteins and the desolvation costs of ligand binding or membrane insertion. However, because of experimental restrictions, absolute solvation free energies of proteins or amino acids are not available. Instead, solvation free energies are estimated based on side chain analog data. This approach implies that the contributions to free energy differences are additive, and it has often been employed for estimating folding or binding free energies. However, it is not clear how much the additivity assumption affects the reliability of the resulting data. Here, we use molecular dynamics-based free energy simulations to calculate absolute hydration free energies for 15 N-acetyl-methylamide amino acids with neutral side chains. By comparing our results with solvation free energies for side chain analogs, we demonstrate that estimates of solvation free energies of full amino acids based on group-additive methods are systematically too negative and completely overestimate the hydrophobicity of glycine. The largest deviation of additive protocols using side chain analog data was 6.7 kcal/mol; on average, the deviation was 4 kcal/mol. We briefly discuss a simple way to alleviate the errors incurred by using side chain analog data and point out the implications of our findings for the field of biophysics and implicit solvent models. To support our results and conclusions, we calculate relative protein stabilities for selected point mutations, yielding a root-mean-square deviation from experimental results of 0.8 kcal/mol.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Hydration free energies of amino acids: why side chain analog data are not enough.J Phys Chem B. 2009 Jul 2;113(26):8967-74. doi: 10.1021/jp902638y. J Phys Chem B. 2009. PMID: 19507836
-
Parameterization of the Hamiltonian Dielectric Solvent (HADES) Reaction-Field Method for the Solvation Free Energies of Amino Acid Side-Chain Analogs.Chemphyschem. 2015 Jun 8;16(8):1739-49. doi: 10.1002/cphc.201402861. Epub 2015 Mar 27. Chemphyschem. 2015. PMID: 25820235
-
Solvation free energies of amino acid side chain analogs for common molecular mechanics water models.J Chem Phys. 2005 Apr 1;122(13):134508. doi: 10.1063/1.1877132. J Chem Phys. 2005. PMID: 15847482
-
Calculation of solvation interaction energies for protein adsorption on polymer surfaces.J Biomater Sci Polym Ed. 1991;3(2):127-47. doi: 10.1163/156856291x00232. J Biomater Sci Polym Ed. 1991. PMID: 1768635 Review.
-
Hydration and heat stability effects on protein unfolding.Prog Biophys Mol Biol. 1993;59(3):237-84. doi: 10.1016/0079-6107(93)90002-2. Prog Biophys Mol Biol. 1993. PMID: 8441810 Review.
Cited by
-
A Heme Propionate Staples the Structure of Cytochrome c for Methionine Ligation to the Heme Iron.Inorg Chem. 2019 Oct 21;58(20):14085-14106. doi: 10.1021/acs.inorgchem.9b02111. Epub 2019 Oct 7. Inorg Chem. 2019. PMID: 31589413 Free PMC article.
-
Peptide Solubility Limits: Backbone and Side-Chain Interactions.J Phys Chem B. 2018 Apr 5;122(13):3528-3539. doi: 10.1021/acs.jpcb.7b10734. Epub 2018 Feb 13. J Phys Chem B. 2018. PMID: 29384681 Free PMC article.
-
Solvation Thermodynamics of Oligoglycine with Respect to Chain Length and Flexibility.Biophys J. 2016 Aug 23;111(4):756-767. doi: 10.1016/j.bpj.2016.07.013. Biophys J. 2016. PMID: 27558719 Free PMC article.
-
A Comparison of QM/MM Simulations with and without the Drude Oscillator Model Based on Hydration Free Energies of Simple Solutes.Molecules. 2018 Oct 19;23(10):2695. doi: 10.3390/molecules23102695. Molecules. 2018. PMID: 30347691 Free PMC article.
-
Importance of Hydrophilic Hydration and Intramolecular Interactions in the Thermodynamics of Helix-Coil Transition and Helix-Helix Assembly in a Deca-Alanine Peptide.J Phys Chem B. 2016 Jan 14;120(1):69-76. doi: 10.1021/acs.jpcb.5b09881. Epub 2015 Dec 22. J Phys Chem B. 2016. PMID: 26649757 Free PMC article.
References
-
- Benson S.W. 2nd Ed. John Wiley; New York: 1976. Thermochemical Kinetics: Methods for Estimation of Thermochemical Data and Rate Parameters.
-
- Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. - PubMed
-
- Tanford C. Contribution of hydrophobic interactions to stability of globular conformation of proteins. J. Am. Chem. Soc. 1962;84:4240–4247.
-
- Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

