Inherent relationships among different biophysical prediction methods for intrinsically disordered proteins
- PMID: 23442871
- PMCID: PMC3552272
- DOI: 10.1016/j.bpj.2012.12.012
Inherent relationships among different biophysical prediction methods for intrinsically disordered proteins
Abstract
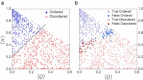
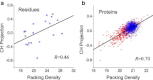
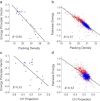
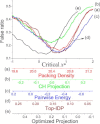
Intrinsically disordered proteins do not have stable secondary and/or tertiary structures but still function. More than 50 prediction methods have been developed and inherent relationships may be expected to exist among them. To investigate this, we conducted molecular simulations and algorithmic analyses on a minimal coarse-grained polypeptide model and discovered a common basis for the charge-hydropathy plot and packing-density algorithms that was verified by correlation analysis. The correlation analysis approach was applied to realistic datasets, which revealed correlations among some physical-chemical properties (charge-hydropathy plot, packing density, pairwise energy). The correlations indicated that these biophysical methods find a projected direction to discriminate ordered and disordered proteins. The optimized projection was determined and the ultimate accuracy limit of the existing algorithms is discussed.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Dunker A.K., Brown C.J., Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. - PubMed
-
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. - PubMed
-
- Huang Y., Liu Z. Intrinsically disordered proteins: the new sequence-structure-function relations. Acta Phys. Chim. Sin. 2010;26:2061–2072.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

