Cell shape can mediate the spatial organization of the bacterial cytoskeleton
- PMID: 23442905
- PMCID: PMC3566457
- DOI: 10.1016/j.bpj.2012.12.027
Cell shape can mediate the spatial organization of the bacterial cytoskeleton
Abstract
The bacterial cytoskeleton guides the synthesis of cell wall and thus regulates cell shape. Because spatial patterning of the bacterial cytoskeleton is critical to the proper control of cell shape, it is important to ask how the cytoskeleton spatially self-organizes in the first place. In this work, we develop a quantitative model to account for the various spatial patterns adopted by bacterial cytoskeletal proteins, especially the orientation and length of cytoskeletal filaments such as FtsZ and MreB in rod-shaped cells. We show that the combined mechanical energy of membrane bending, membrane pinning, and filament bending of a membrane-attached cytoskeletal filament can be sufficient to prescribe orientation, e.g., circumferential for FtsZ or helical for MreB, with the accuracy of orientation increasing with the length of the cytoskeletal filament. Moreover, the mechanical energy can compete with the chemical energy of cytoskeletal polymerization to regulate filament length. Notably, we predict a conformational transition with increasing polymer length from smoothly curved to end-bent polymers. Finally, the mechanical energy also results in a mutual attraction among polymers on the same membrane, which could facilitate tight polymer spacing or bundling. The predictions of the model can be verified through genetic, microscopic, and microfluidic approaches.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


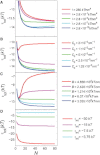

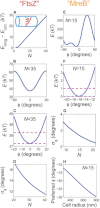

References
-
- Cooper S., Denny M.W. A conjecture on the relationship of bacterial shape to motility in rod-shaped bacteria. FEMS Microbiol. Lett. 1997;148:227–231.
-
- Cabeen M.T., Jacobs-Wagner C. Bacterial cell shape. Nat. Rev. Microbiol. 2005;3:601–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

