Computational assembly of polymorphic amyloid fibrils reveals stable aggregates
- PMID: 23442919
- PMCID: PMC3566450
- DOI: 10.1016/j.bpj.2012.12.037
Computational assembly of polymorphic amyloid fibrils reveals stable aggregates
Abstract
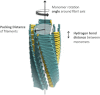
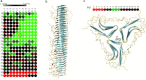
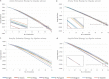
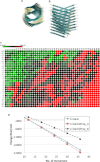
Amyloid proteins aggregate into polymorphic fibrils that damage tissues of the brain, nerves, and heart. Experimental and computational studies have examined the structural basis and the nucleation of short fibrils, but the ability to predict and precisely quantify the stability of larger aggregates has remained elusive. We established a complete classification of fibril shapes and developed a tool called CreateFibril to build such complex, polymorphic, modular structures automatically. We applied stability landscapes, a technique we developed to reveal reliable fibril structural parameters, to assess fibril stability. CreateFibril constructed HET-s, Aβ, and amylin fibrils up to 17 nm in length, and utilized a novel dipolar solvent model that captured the effect of dipole-dipole interactions between water and very large molecular systems to assess their aqueous stability. Our results validate experimental data for HET-s and Aβ, and suggest novel (to our knowledge) findings for amylin. In particular, we predicted the correct structural parameters (rotation angles, packing distances, hydrogen bond lengths, and helical pitches) for the one and three predominant HET-s protofilaments. We reveal and structurally characterize all known Aβ polymorphic fibrils, including structures recently classified as wrapped fibrils. Finally, we elucidate the predominant amylin fibrils and assert that native amylin is more stable than its amyloid form. CreateFibril and a database of all stable polymorphic fibril models we tested, along with their structural energy landscapes, are available at http://amyloid.cs.mcgill.ca.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Heterogeneous triangular structures of human islet amyloid polypeptide (amylin) with internal hydrophobic cavity and external wrapping morphology reveal the polymorphic nature of amyloid fibrils.Biomacromolecules. 2011 May 9;12(5):1781-94. doi: 10.1021/bm2001507. Epub 2011 Apr 5. Biomacromolecules. 2011. PMID: 21428404
-
Polymorphic structures of Alzheimer's β-amyloid globulomers.PLoS One. 2011;6(6):e20575. doi: 10.1371/journal.pone.0020575. Epub 2011 Jun 7. PLoS One. 2011. PMID: 21687730 Free PMC article.
-
Computational re-engineering of Amylin sequence with reduced amyloidogenic potential.BMC Struct Biol. 2015 Apr 24;15:7. doi: 10.1186/s12900-015-0034-4. BMC Struct Biol. 2015. PMID: 25903685 Free PMC article.
-
Probing the pressure-temperature stability of amyloid fibrils provides new insights into their molecular properties.Biochim Biophys Acta. 2006 Mar;1764(3):452-60. doi: 10.1016/j.bbapap.2005.10.021. Epub 2005 Nov 16. Biochim Biophys Acta. 2006. PMID: 16337233 Review.
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
Cited by
-
Exploring Abeta42 monomer diffusion dynamics on fibril surfaces through molecular simulations.Protein Sci. 2025 Jun;34(6):e70131. doi: 10.1002/pro.70131. Protein Sci. 2025. PMID: 40371804
-
On the lack of polymorphism in Aβ-peptide aggregates derived from patient brains.Protein Sci. 2015 Jun;24(6):923-35. doi: 10.1002/pro.2668. Epub 2015 Apr 14. Protein Sci. 2015. PMID: 25739352 Free PMC article.
-
Probing the binding affinity of amyloids to reduce toxicity of oligomers in diabetes.Bioinformatics. 2015 Jul 15;31(14):2294-302. doi: 10.1093/bioinformatics/btv143. Epub 2015 Mar 15. Bioinformatics. 2015. PMID: 25777526 Free PMC article.
-
Inter-species cross-seeding: stability and assembly of rat-human amylin aggregates.PLoS One. 2014 May 8;9(5):e97051. doi: 10.1371/journal.pone.0097051. eCollection 2014. PLoS One. 2014. PMID: 24810618 Free PMC article.
-
Diabetes Drug Discovery: hIAPP1-37 Polymorphic Amyloid Structures as Novel Therapeutic Targets.Molecules. 2018 Mar 19;23(3):686. doi: 10.3390/molecules23030686. Molecules. 2018. PMID: 29562662 Free PMC article.
References
-
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. - PubMed
-
- Uversky V.N., Fink A.L. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. - PubMed
-
- Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. - PubMed
-
- Kirkwood S.C., Su J.L., Foroud T. Progression of symptoms in the early and middle stages of Huntington disease. Arch. Neurol. 2001;58:273–278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

