H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis
- PMID: 23443005
- PMCID: PMC3585446
- DOI: 10.1128/mBio.00062-13
H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis
Abstract
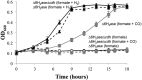
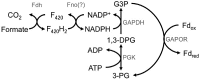
Hydrogenotrophic methanogenic Archaea require reduced ferredoxin as an anaplerotic source of electrons for methanogenesis. H(2) oxidation by the hydrogenase Eha provides these electrons, consistent with an H(2) requirement for growth. Here we report the identification of alternative pathways of ferredoxin reduction in Methanococcus maripaludis that operate independently of Eha to stimulate methanogenesis. A suppressor mutation that increased expression of the glycolytic enzyme glyceraldehyde-3-phosphate:ferredoxin oxidoreductase resulted in a strain capable of H(2)-independent ferredoxin reduction and growth with formate as the sole electron donor. In this background, it was possible to eliminate all seven hydrogenases of M. maripaludis. Alternatively, carbon monoxide oxidation by carbon monoxide dehydrogenase could also generate reduced ferredoxin that feeds into methanogenesis. In either case, the reduced ferredoxin generated was inefficient at stimulating methanogenesis, resulting in a slow growth phenotype. As methanogenesis is limited by the availability of reduced ferredoxin under these conditions, other electron donors, such as reduced coenzyme F(420), should be abundant. Indeed, when F(420)-reducing hydrogenase was reintroduced into the hydrogenase-free mutant, the equilibrium of H(2) production via an F(420)-dependent formate:H(2) lyase activity shifted markedly toward H(2) compared to the wild type.
Importance: Hydrogenotrophic methanogens are thought to require H(2) as a substrate for growth and methanogenesis. Here we show alternative pathways in methanogenic metabolism that alleviate this H(2) requirement and demonstrate, for the first time, a hydrogenotrophic methanogen that is capable of growth in the complete absence of H(2). The demonstration of alternative pathways in methanogenic metabolism suggests that this important group of organisms is metabolically more versatile than previously thought.
Figures




Similar articles
-
Effects of H2 and formate on growth yield and regulation of methanogenesis in Methanococcus maripaludis.J Bacteriol. 2013 Apr;195(7):1456-62. doi: 10.1128/JB.02141-12. Epub 2013 Jan 18. J Bacteriol. 2013. PMID: 23335420 Free PMC article.
-
Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis.Appl Environ Microbiol. 2008 Nov;74(21):6584-90. doi: 10.1128/AEM.01455-08. Epub 2008 Sep 12. Appl Environ Microbiol. 2008. PMID: 18791018 Free PMC article.
-
Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis.Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15473-8. doi: 10.1073/pnas.1208779109. Epub 2012 Aug 7. Proc Natl Acad Sci U S A. 2012. PMID: 22872868 Free PMC article.
-
Metabolic processes of Methanococcus maripaludis and potential applications.Microb Cell Fact. 2016 Jun 10;15(1):107. doi: 10.1186/s12934-016-0500-0. Microb Cell Fact. 2016. PMID: 27286964 Free PMC article. Review.
-
Energy Conservation and Hydrogenase Function in Methanogenic Archaea, in Particular the Genus Methanosarcina.Microbiol Mol Biol Rev. 2019 Sep 18;83(4):e00020-19. doi: 10.1128/MMBR.00020-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31533962 Free PMC article. Review.
Cited by
-
Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation.Microbiol Mol Biol Rev. 2014 Mar;78(1):89-175. doi: 10.1128/MMBR.00041-13. Microbiol Mol Biol Rev. 2014. PMID: 24600042 Free PMC article. Review.
-
Hydrogenase-independent uptake and metabolism of electrons by the archaeon Methanococcus maripaludis.ISME J. 2014 Aug;8(8):1673-81. doi: 10.1038/ismej.2014.82. Epub 2014 May 20. ISME J. 2014. PMID: 24844759 Free PMC article.
-
Effects of isoleucine 135 side chain length on the cofactor donor-acceptor distance within F420H2:NADP+ oxidoreductase: A kinetic analysis.Biochem Biophys Rep. 2016 Nov 30;9:114-120. doi: 10.1016/j.bbrep.2016.11.012. eCollection 2017 Mar. Biochem Biophys Rep. 2016. PMID: 28955995 Free PMC article.
-
Biogeochemical Cycling by a Low-Diversity Microbial Community in Deep Groundwater.Front Microbiol. 2018 Sep 7;9:2129. doi: 10.3389/fmicb.2018.02129. eCollection 2018. Front Microbiol. 2018. PMID: 30245678 Free PMC article.
-
Exploring Hydrogenotrophic Methanogenesis: a Genome Scale Metabolic Reconstruction of Methanococcus maripaludis.J Bacteriol. 2016 Nov 18;198(24):3379-3390. doi: 10.1128/JB.00571-16. Print 2016 Dec 15. J Bacteriol. 2016. PMID: 27736793 Free PMC article.
References
-
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579–591 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
