In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion
- PMID: 23443137
- PMCID: PMC3675817
- DOI: 10.1074/mcp.M112.022848
In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion
Abstract
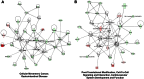
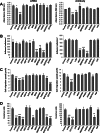
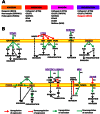
Liver metastasis in colorectal cancer is the major cause of cancer-related deaths. To identify and characterize proteins associated with colon cancer metastasis, we have compared the conditioned serum-free medium of highly metastatic KM12SM colorectal cancer cells with the parental, poorly metastatic KM12C cells using quantitative stable isotope labeling by amino acids in cell culture (SILAC) analyses on a linear ion trap-Orbitrap Velos mass spectrometer. In total, 1337 proteins were simultaneously identified in SILAC forward and reverse experiments. For quantification, 1098 proteins were selected in both experiments, with 155 proteins showing >1.5-fold change. About 52% of these proteins were secreted directly or using alternative secretion pathways. GDF15, S100A8/A9, and SERPINI1 showed capacity to discriminate cancer serum samples from healthy controls using ELISAs. In silico analyses of deregulated proteins in the secretome of metastatic cells showed a major abundance of proteins involved in cell adhesion, migration, and invasion. To characterize the tumorigenic and metastatic properties of some top up- and down-regulated proteins, we used siRNA silencing and antibody blocking. Knockdown expression of NEO1, SERPINI1, and PODXL showed a significant effect on cellular adhesion. Silencing or blocking experiments with SOSTDC1, CTSS, EFNA3, CD137L/TNFSF9, ZG16B, and Midkine caused a significant decrease in migration and invasion of highly metastatic cells. In addition, silencing of SOSTDC1, EFNA3, and CD137L/TNFSF9 reduced liver colonization capacity of KM12SM cells. Finally, the panel of six proteins involved in invasion showed association with poor prognosis and overall survival after dataset analysis of gene alterations. In summary, we have defined a collection of proteins that are relevant for understanding the mechanisms underlying adhesion, migration, invasion, and metastasis in colorectal cancer.
Figures






References
-
- Locker G. Y., Hamilton S., Harris J., Jessup J. M., Kemeny N., Macdonald J. S., Somerfield M. R., Hayes D. F., Bast R. C., Jr. (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 24, 5313–5327 - PubMed
-
- Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43, 1348–1360 - PubMed
-
- Babel I., Barderas R., Diaz-Uriarte R., Moreno V., Suarez A., Fernandez-Acenero M. J., Salazar R., Capella G., Casal J. I. (2011) Identification of MST1/STK4 and SULF1 proteins as autoantibody targets for the diagnosis of colorectal cancer by using phage microarrays. Mol. Cell. Proteomics 10, M110.001784. - PMC - PubMed
-
- Hundt S., Haug U., Brenner H. (2007) Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol. Biomarkers Prev. 16, 1935–1953 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

