Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella
- PMID: 23443158
- PMCID: PMC3634460
- DOI: 10.3390/ijms14034560
Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella
Abstract
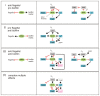
Biofilm formation in Escherichia coli and other enteric bacteria involves the inverse regulation of the synthesis of flagella and biofilm matrix components such as amyloid curli fibres, cellulose, colanic acid and poly-N-acetylglucosamine (PGA). Physiologically, these processes reflect the transition from growth to stationary phase. At the molecular level, they are tightly controlled by various sigma factors competing for RNA polymerase, a series of transcription factors acting in hierarchical regulatory cascades and several nucleotide messengers, including cyclic-di-GMP. In addition, a surprisingly large number of small regulatory RNAs (sRNAs) have been shown to directly or indirectly modulate motility and/or biofilm formation. This review aims at giving an overview of these sRNA regulators and their impact in biofilm formation in E. coli and Salmonella. Special emphasis will be put on sRNAs, that have known targets such as the mRNAs of the flagellar master regulator FlhDC, the stationary phase sigma factor σS (RpoS) and the key biofilm regulator CsgD that have recently been shown to act as major hubs for regulation by multiple sRNAs.
Figures


References
-
- Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. - PubMed
-
- Branda S.S., Vik S., Friedman L., Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13:20–26. - PubMed
-
- Anderson G.G., O’Toole G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008;322:85–105. - PubMed
-
- Danese P.N., Pratt L.A., Dove S.L., Kolter R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 2000;37:424–432. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

