Clonal diversity of recurrently mutated genes in myelodysplastic syndromes
- PMID: 23443460
- PMCID: PMC3736571
- DOI: 10.1038/leu.2013.58
Clonal diversity of recurrently mutated genes in myelodysplastic syndromes
Abstract
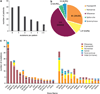
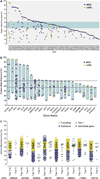
Recent studies suggest that most cases of myelodysplastic syndrome (MDS) are clonally heterogeneous, with a founding clone and multiple subclones. It is not known whether specific gene mutations typically occur in founding clones or subclones. We screened a panel of 94 candidate genes in a cohort of 157 patients with MDS or secondary acute myeloid leukemia (sAML). This included 150 cases with samples obtained at MDS diagnosis and 15 cases with samples obtained at sAML transformation (8 were also analyzed at the MDS stage). We performed whole-genome sequencing (WGS) to define the clonal architecture in eight sAML genomes and identified the range of variant allele frequencies (VAFs) for founding clone mutations. At least one mutation or cytogenetic abnormality was detected in 83% of the 150 MDS patients and 17 genes were significantly mutated (false discovery rate ≤0.05). Individual genes and patient samples displayed a wide range of VAFs for recurrently mutated genes, indicating that no single gene is exclusively mutated in the founding clone. The VAFs of recurrently mutated genes did not fully recapitulate the clonal architecture defined by WGS, suggesting that comprehensive sequencing may be required to accurately assess the clonal status of recurrently mutated genes in MDS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Understanding the mutational evolution of clonal cytopenias and oligoblastic myelogenous leukemia ('myelodysplasia').Leukemia. 2014 Jan;28(1):202. doi: 10.1038/leu.2013.209. Epub 2013 Jul 11. Leukemia. 2014. PMID: 23842426 No abstract available.
References
-
- Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. - PubMed
-
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. - PubMed
Publication types
MeSH terms
Grants and funding
- P01CA101937/CA/NCI NIH HHS/United States
- R01 HL082973/HL/NHLBI NIH HHS/United States
- RC2 HL102927/HL/NHLBI NIH HHS/United States
- U01 HG006517/HG/NHGRI NIH HHS/United States
- R01HL082973/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- P01 CA101937/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- RC2HL102927/HL/NHLBI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U01HG006517/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

