Genome maintenance and transcription integrity in aging and disease
- PMID: 23443494
- PMCID: PMC3580961
- DOI: 10.3389/fgene.2013.00019
Genome maintenance and transcription integrity in aging and disease
Abstract
DNA damage contributes to cancer development and aging. Congenital syndromes that affect DNA repair processes are characterized by cancer susceptibility, developmental defects, and accelerated aging (Schumacher et al., 2008). DNA damage interferes with DNA metabolism by blocking replication and transcription. DNA polymerase blockage leads to replication arrest and can gives rise to genome instability. Transcription, on the other hand, is an essential process for utilizing the information encoded in the genome. DNA damage that interferes with transcription can lead to apoptosis and cellular senescence. Both processes are powerful tumor suppressors (Bartek and Lukas, 2007). Cellular response mechanisms to stalled RNA polymerase II complexes have only recently started to be uncovered. Transcription-coupled DNA damage responses might thus play important roles for the adjustments to DNA damage accumulation in the aging organism (Garinis et al., 2009). Here we review human disorders that are caused by defects in genome stability to explore the role of DNA damage in aging and disease. We discuss how the nucleotide excision repair system functions at the interface of transcription and repair and conclude with concepts how therapeutic targeting of transcription might be utilized in the treatment of cancer.
Keywords: DNA damage; DNA repair; cancer; genetic; progeria; transcription.
Figures
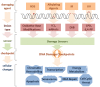
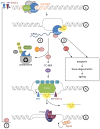
References
-
- Anindya R., Aygun O., Svejstrup J. Q. (2007). Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell 28 386–397 - PubMed
-
- Bartek J., Lukas J. (2007). DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19 238–245 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

