Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum
- PMID: 23443558
- PMCID: PMC3586713
- DOI: 10.1038/ncomms2539
Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum
Abstract
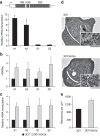
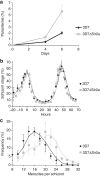
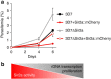
The Plasmodium falciparum histone deacetylase Sir2a localizes at telomeric regions where it contributes to epigenetic silencing of clonally variant virulence genes. Apart from telomeres, PfSir2a also accumulates in the nucleolus, which harbours the developmentally regulated ribosomal RNA genes. Here we investigate the nucleolar function of PfSir2a and demonstrate that PfSir2a fine-tunes ribosomal RNA gene transcription. Using a parasite line in which PfSir2a has been disrupted, we observe that histones near the transcription start sites of all ribosomal RNA genes are hyperacetylated and that transcription of ribosomal RNA genes is upregulated. Complementation of the PfSir2a-disrupted parasites restores the ribosomal RNA levels, whereas PfSir2a overexpression in wild-type parasites decreases ribosomal RNA synthesis. Furthermore, we observe that PfSir2a modulation of ribosomal RNA synthesis is linked to an altered number of daughter merozoites and the parasite multiplication rate. These findings provide new insights into an epigenetic mechanism that controls malaria parasite proliferation and virulence.
Figures

Similar articles
-
Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter.PLoS Pathog. 2011 Feb;7(2):e1001292. doi: 10.1371/journal.ppat.1001292. Epub 2011 Feb 17. PLoS Pathog. 2011. PMID: 21379342 Free PMC article.
-
The effect of Plasmodium falciparum Sir2a histone deacetylase on clonal and longitudinal variation in expression of the var family of virulence genes.Int J Parasitol. 2010 Jan;40(1):35-43. doi: 10.1016/j.ijpara.2009.06.012. Epub 2009 Aug 8. Int J Parasitol. 2010. PMID: 19666023
-
Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum.PLoS Biol. 2009 Apr 14;7(4):e84. doi: 10.1371/journal.pbio.1000084. PLoS Biol. 2009. PMID: 19402747 Free PMC article.
-
High resolution 3D perspective of Plasmodium biology: advancing into a new era.Trends Parasitol. 2011 Dec;27(12):548-54. doi: 10.1016/j.pt.2011.08.002. Epub 2011 Sep 4. Trends Parasitol. 2011. PMID: 21893431 Review.
-
Host cell invasion by malaria parasites.Parasitol Today. 2000 Oct;16(10):411-5. doi: 10.1016/s0169-4758(00)01756-7. Parasitol Today. 2000. PMID: 11006471 Review.
Cited by
-
Heat Shock Protein 90 Regulates the Activity of Histone Deacetylase Sir2 in Plasmodium falciparum.mSphere. 2022 Oct 26;7(5):e0032922. doi: 10.1128/msphere.00329-22. Epub 2022 Sep 19. mSphere. 2022. PMID: 36121150 Free PMC article.
-
Hierarchical transcriptional control regulates Plasmodium falciparum sexual differentiation.BMC Genomics. 2019 Dec 3;20(1):920. doi: 10.1186/s12864-019-6322-9. BMC Genomics. 2019. PMID: 31795940 Free PMC article.
-
Hungry for control: metabolite signaling to chromatin in Plasmodium falciparum.Curr Opin Microbiol. 2024 Apr;78:102430. doi: 10.1016/j.mib.2024.102430. Epub 2024 Feb 2. Curr Opin Microbiol. 2024. PMID: 38306915 Free PMC article. Review.
-
Adaptation of Plasmodium falciparum to its transmission environment.Nat Ecol Evol. 2018 Feb;2(2):377-387. doi: 10.1038/s41559-017-0419-9. Epub 2017 Dec 18. Nat Ecol Evol. 2018. PMID: 29255304 Free PMC article.
-
How Many Is Enough? - Challenges of Multinucleated Cell Division in Malaria Parasites.Front Cell Infect Microbiol. 2021 May 7;11:658616. doi: 10.3389/fcimb.2021.658616. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34026661 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

