Coactivators enable glucocorticoid receptor recruitment to fine-tune estrogen receptor transcriptional responses
- PMID: 23444138
- PMCID: PMC3627592
- DOI: 10.1093/nar/gkt100
Coactivators enable glucocorticoid receptor recruitment to fine-tune estrogen receptor transcriptional responses
Abstract
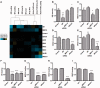
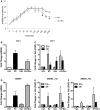
Nuclear receptors (NRs) are central regulators of pathophysiological processes; however, how their responses intertwine is still not fully understood. The aim of this study was to determine whether and how steroid NRs can influence each other's activity under co-agonist treatment. We used a unique system consisting of a multicopy integration of an estrogen receptor responsive unit that allows direct visualization and quantification of estrogen receptor alpha (ERα) DNA binding, co-regulator recruitment and transcriptional readout. We find that ERα DNA loading is required for other type I nuclear receptors to be co-recruited after dual agonist treatment. We focused on ERα/glucocorticoid receptor interplay and demonstrated that it requires steroid receptor coactivators (SRC-2, SRC-3) and the mediator component MED14. We then validated this cooperative interplay on endogenous target genes in breast cancer cells. Taken together, this work highlights another layer of mechanistic complexity through which NRs cross-talk with each other on chromatin under multiple hormonal stimuli.
Figures
References
-
- Horner-Glister E, Maleki-Dizaji M, Guerin CJ, Johnson SM, Styles J, White IN. Influence of oestradiol and tamoxifen on oestrogen receptors-alpha and -beta protein degradation and non-genomic signalling pathways in uterine and breast carcinoma cells. J. Mol. Endocrinol. 2005;35:421–432. - PubMed
-
- Ogawa S, Eng V, Taylor J, Lubahn DB, Korach KS, Pfaff DW. Roles of estrogen receptor-alpha gene expression in reproduction-related behaviors in female mice. Endocrinology. 1998;139:5070–5081. - PubMed
-
- Rudnik V, Sanyal A, Syed FA, Monroe DG, Spelsberg TC, Oursler MJ, Khosla S. Loss of ERE binding activity by estrogen receptor-alpha alters basal and estrogen-stimulated bone-related gene expression by osteoblastic cells. J. Cell. Biochem. 2008;103:896–907. - PubMed
-
- Tobias JH, Steer CD, Vilarino-Guell C, Brown MA. Estrogen receptor alpha regulates area-adjusted bone mineral content in late pubertal girls. J. Clin. Endocrinol. Metab. 2007;92:641–647. - PubMed
-
- Pike AC. Lessons learnt from structural studies of the oestrogen receptor. Best Pract. Res. 2006;20:1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

