CoRSeqV3-C: a novel HIV-1 subtype C specific V3 sequence based coreceptor usage prediction algorithm
- PMID: 23446039
- PMCID: PMC3599735
- DOI: 10.1186/1742-4690-10-24
CoRSeqV3-C: a novel HIV-1 subtype C specific V3 sequence based coreceptor usage prediction algorithm
Abstract
Background: The majority of HIV-1 subjects worldwide are infected with HIV-1 subtype C (C-HIV). Although C-HIV predominates in developing regions of the world such as Southern Africa and Central Asia, C-HIV is also spreading rapidly in countries with more developed economies and health care systems, whose populations are more likely to have access to wider treatment options, including the CCR5 antagonist maraviroc (MVC). The ability to reliably determine C-HIV coreceptor usage is therefore becoming increasingly more important. In silico V3 sequence based coreceptor usage prediction algorithms are a relatively rapid and cost effective method for determining HIV-1 coreceptor specificity. In this study, we elucidated the V3 sequence determinants of C-HIV coreceptor usage, and used this knowledge to develop and validate a novel, user friendly, and highly sensitive C-HIV specific coreceptor usage prediction algorithm.
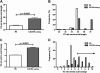
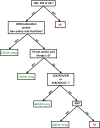
Results: We characterized every phenotypically-verified C-HIV gp120 V3 sequence available in the Los Alamos HIV Database. Sequence analyses revealed that compared to R5 C-HIV V3 sequences, CXCR4-using C-HIV V3 sequences have significantly greater amino acid variability, increased net charge, increased amino acid length, increased frequency of insertions and substitutions within the GPGQ crown motif, and reduced frequency of glycosylation sites. Based on these findings, we developed a novel C-HIV specific coreceptor usage prediction algorithm (CoRSeqV3-C), which we show has superior sensitivity for determining CXCR4 usage by C-HIV strains compared to all other available algorithms and prediction rules, including Geno2pheno[coreceptor] and WebPSSMSINSI-C, which has been designed specifically for C-HIV.
Conclusions: CoRSeqV3-C is now openly available for public use at http://www.burnet.edu.au/coreceptor. Our results show that CoRSeqV3-C is the most sensitive V3 sequence based algorithm presently available for predicting CXCR4 usage of C-HIV strains, without compromising specificity. CoRSeqV3-C may be potentially useful for assisting clinicians to decide the best treatment options for patients with C-HIV infection, and will be helpful for basic studies of C-HIV pathogenesis.
Figures
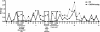
Similar articles
-
Appraising the performance of genotyping tools in the prediction of coreceptor tropism in HIV-1 subtype C viruses.BMC Infect Dis. 2012 Sep 2;12:203. doi: 10.1186/1471-2334-12-203. BMC Infect Dis. 2012. PMID: 22938574 Free PMC article.
-
Covariance of charged amino acids at positions 322 and 440 of HIV-1 Env contributes to coreceptor specificity of subtype B viruses, and can be used to improve the performance of V3 sequence-based coreceptor usage prediction algorithms.PLoS One. 2014 Oct 14;9(10):e109771. doi: 10.1371/journal.pone.0109771. eCollection 2014. PLoS One. 2014. PMID: 25313689 Free PMC article.
-
Performance of genotypic tools for prediction of tropism in HIV-1 subtype C V3 loop sequences.Intervirology. 2015;58(1):1-5. doi: 10.1159/000369017. Epub 2015 Jan 7. Intervirology. 2015. PMID: 25573618
-
HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis.AIDS Res Ther. 2020 Feb 7;17(1):5. doi: 10.1186/s12981-020-0263-x. AIDS Res Ther. 2020. PMID: 32033571 Free PMC article. Review.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
Cited by
-
Longitudinal analysis of subtype C envelope tropism for memory CD4+ T cell subsets over the first 3 years of untreated HIV-1 infection.Retrovirology. 2020 Aug 6;17(1):24. doi: 10.1186/s12977-020-00532-2. Retrovirology. 2020. PMID: 32762760 Free PMC article.
-
The Molecular Basis of pH-Modulated HIV gp120 Binding Revealed.Evol Bioinform Online. 2019 Mar 7;15:1176934319831308. doi: 10.1177/1176934319831308. eCollection 2019. Evol Bioinform Online. 2019. PMID: 30872918 Free PMC article.
-
Novel Strategy To Adapt Simian-Human Immunodeficiency Virus E1 Carrying env from an RV144 Volunteer to Rhesus Macaques: Coreceptor Switch and Final Recovery of a Pathogenic Virus with Exclusive R5 Tropism.J Virol. 2018 Jun 29;92(14):e02222-17. doi: 10.1128/JVI.02222-17. Print 2018 Jul 15. J Virol. 2018. PMID: 29743361 Free PMC article.
-
Coreceptor usage, diversity, and divergence in drug-naive and drug-exposed individuals from Malawi, infected with HIV-1 subtype C for more than 20 years.AIDS Res Hum Retroviruses. 2014 Oct;30(10):975-83. doi: 10.1089/aid.2013.0240. Epub 2014 Jul 29. AIDS Res Hum Retroviruses. 2014. PMID: 24925099 Free PMC article.
-
Reliable genotypic tropism tests for the major HIV-1 subtypes.Sci Rep. 2015 Feb 25;5:8543. doi: 10.1038/srep08543. Sci Rep. 2015. PMID: 25712827 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

