The role of the nucleosome acidic patch in modulating higher order chromatin structure
- PMID: 23446052
- PMCID: PMC3627075
- DOI: 10.1098/rsif.2012.1022
The role of the nucleosome acidic patch in modulating higher order chromatin structure
Abstract
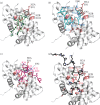
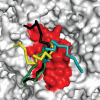
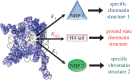
Higher order folding of chromatin fibre is mediated by interactions of the histone H4 N-terminal tail domains with neighbouring nucleosomes. Mechanistically, the H4 tails of one nucleosome bind to the acidic patch region on the surface of adjacent nucleosomes, causing fibre compaction. The functionality of the chromatin fibre can be modified by proteins that interact with the nucleosome. The co-structures of five different proteins with the nucleosome (LANA, IL-33, RCC1, Sir3 and HMGN2) recently have been examined by experimental and computational studies. Interestingly, each of these proteins displays steric, ionic and hydrogen bond complementarity with the acidic patch, and therefore will compete with each other for binding to the nucleosome. We first review the molecular details of each interface, focusing on the key non-covalent interactions that stabilize the protein-acidic patch interactions. We then propose a model in which binding of proteins to the nucleosome disrupts interaction of the H4 tail domains with the acidic patch, preventing the intrinsic chromatin folding pathway and leading to assembly of alternative higher order chromatin structures with unique biological functions.
Figures


Similar articles
-
Chromatin modification by PSC occurs at one PSC per nucleosome and does not require the acidic patch of histone H2A.PLoS One. 2012;7(10):e47162. doi: 10.1371/journal.pone.0047162. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071745 Free PMC article.
-
Regulation of Nucleosome Stacking and Chromatin Compaction by the Histone H4 N-Terminal Tail-H2A Acidic Patch Interaction.J Mol Biol. 2017 Jun 30;429(13):2075-2092. doi: 10.1016/j.jmb.2017.03.016. Epub 2017 Mar 16. J Mol Biol. 2017. PMID: 28322915
-
Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome.J Mol Biol. 2012 Aug 3;421(1):30-7. doi: 10.1016/j.jmb.2012.04.032. Epub 2012 May 7. J Mol Biol. 2012. PMID: 22575889 Free PMC article.
-
Targeting the Nucleosome Acidic Patch by Viral Proteins: Two Birds with One Stone?mBio. 2022 Apr 26;13(2):e0173321. doi: 10.1128/mbio.01733-21. Epub 2022 Mar 28. mBio. 2022. PMID: 35343785 Free PMC article. Review.
-
Beyond the Nucleosome: Nucleosome-Protein Interactions and Higher Order Chromatin Structure.J Mol Biol. 2021 Mar 19;433(6):166827. doi: 10.1016/j.jmb.2021.166827. Epub 2021 Jan 16. J Mol Biol. 2021. PMID: 33460684 Review.
Cited by
-
Nucleosome allostery in pioneer transcription factor binding.Proc Natl Acad Sci U S A. 2020 Aug 25;117(34):20586-20596. doi: 10.1073/pnas.2005500117. Epub 2020 Aug 10. Proc Natl Acad Sci U S A. 2020. PMID: 32778600 Free PMC article.
-
The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in Saccharomyces cerevisiae.PLoS Genet. 2015 Aug 4;11(8):e1005420. doi: 10.1371/journal.pgen.1005420. eCollection 2015 Aug. PLoS Genet. 2015. PMID: 26241481 Free PMC article.
-
The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling.Nat Commun. 2018 Aug 17;9(1):3309. doi: 10.1038/s41467-018-05710-7. Nat Commun. 2018. PMID: 30120252 Free PMC article.
-
The solid and liquid states of chromatin.Epigenetics Chromatin. 2021 Oct 30;14(1):50. doi: 10.1186/s13072-021-00424-5. Epigenetics Chromatin. 2021. PMID: 34717733 Free PMC article. Review.
-
Critical Roles of IL-33/ST2 Pathway in Neurological Disorders.Mediators Inflamm. 2018 Feb 4;2018:5346413. doi: 10.1155/2018/5346413. eCollection 2018. Mediators Inflamm. 2018. PMID: 29507527 Free PMC article. Review.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 7, 251–260 - PubMed
-
- Hansen JC. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31, 361–39210.1146/annurev.biophys.31.101101.140858 (doi:10.1146/annurev.biophys.31.101101.140858) - DOI - DOI - PubMed
-
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–9610.1016/S0022-2836(03)00025-1 (doi:10.1016/S0022-2836(03)00025-1) - DOI - DOI - PubMed
-
- Gordon F, Luger K, Hansen JC. 2005. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 280, 33 701–33 70610.1074/jbc.M507048200 (doi:10.1074/jbc.M507048200) - DOI - DOI - PubMed
-
- McBryant S, Klonoski J, Sorensen TC, Norskog SS, Williams S, Resch MG, Toombs JA, Hobdey SE, Hansen JC. 2009. Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization: roles of amino acid sequence, domain length, and charge density. J. Biol. Chem. 25, 16 716–16 72210.1074/jbc.M109.011288 (doi:10.1074/jbc.M109.011288) - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

