Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data
- PMID: 23446317
- PMCID: PMC3599118
- DOI: 10.1186/1471-2334-13-110
Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data
Abstract
Background: Mycobacterium tuberculosis is characterised by limited genomic diversity, which makes the application of whole genome sequencing particularly attractive for clinical and epidemiological investigation. However, in order to confidently infer transmission events, an accurate knowledge of the rate of change in the genome over relevant timescales is required.
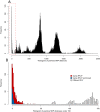
Methods: We attempted to estimate a molecular clock by sequencing 199 isolates from epidemiologically linked tuberculosis cases, collected in the Netherlands spanning almost 16 years.
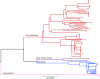
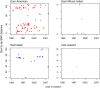
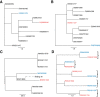
Results: Multiple analyses support an average mutation rate of ~0.3 SNPs per genome per year. However, all analyses revealed a very high degree of variation around this mean, making the confirmation of links proposed by epidemiology, and inference of novel links, difficult. Despite this, in some cases, the phylogenetic context of other strains provided evidence supporting the confident exclusion of previously inferred epidemiological links.
Conclusions: This in-depth analysis of the molecular clock revealed that it is slow and variable over short time scales, which limits its usefulness in transmission studies. However, the superior resolution of whole genome sequencing can provide the phylogenetic context to allow the confident exclusion of possible transmission events previously inferred via traditional DNA fingerprinting techniques and epidemiological cluster investigation. Despite the slow generation of variation even at the whole genome level we conclude that the investigation of tuberculosis transmission will benefit greatly from routine whole genome sequencing.
Figures



References
-
- WHO. Global tuberculosis report 2012. Geneva, Switzerland: World Health Organisation, WHO Press; 2012.
-
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of mycobacterium tuberculosis. J Clin Microbiol. 2006;44(12):4498–4510. doi: 10.1128/JCM.01392-06. - DOI - PMC - PubMed
-
- Niemann S, Koser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA. Genomic diversity among drug sensitive and multidrug resistant isolates of mycobacterium tuberculosis with identical DNA fingerprints. PLoS One. 2009;4(10):e7407. doi: 10.1371/journal.pone.0007407. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

