Structural visualization of key steps in human transcription initiation
- PMID: 23446344
- PMCID: PMC3612373
- DOI: 10.1038/nature11991
Structural visualization of key steps in human transcription initiation
Abstract
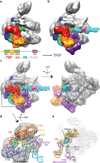
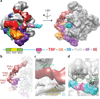
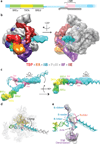
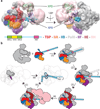
Eukaryotic transcription initiation requires the assembly of general transcription factors into a pre-initiation complex that ensures the accurate loading of RNA polymerase II (Pol II) at the transcription start site. The molecular mechanism and function of this assembly have remained elusive due to lack of structural information. Here we have used an in vitro reconstituted system to study the stepwise assembly of human TBP, TFIIA, TFIIB, Pol II, TFIIF, TFIIE and TFIIH onto promoter DNA using cryo-electron microscopy. Our structural analyses provide pseudo-atomic models at various stages of transcription initiation that illuminate critical molecular interactions, including how TFIIF engages Pol II and promoter DNA to stabilize both the closed pre-initiation complex and the open-promoter complex, and to regulate start--initiation complexes, combined with the localization of the TFIIH helicases XPD and XPB, support a DNA translocation model of XPB and explain its essential role in promoter opening.
Figures

References
-
- Matsui T, Segall J, Weil PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980;255:11992–11996. - PubMed
-
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. - PubMed
-
- Goodrich JA, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. - PubMed
-
- Cramer P, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

