Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells
- PMID: 23447566
- PMCID: PMC3607057
- DOI: 10.1073/pnas.1300392110
Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells
Abstract
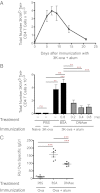
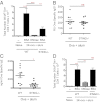
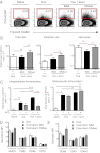
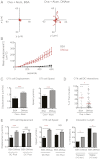
Many vaccines include aluminum salts (alum) as adjuvants despite little knowledge of alum's functions. Host DNA rapidly coats injected alum. Here, we further investigated the mechanism of alum and DNA's adjuvant function. Our data show that DNase coinjection reduces CD4 T-cell priming by i.m. injected antigen + alum. This effect is partially replicated in mice lacking stimulator of IFN genes, a mediator of cellular responses to cytoplasmic DNA. Others have shown that DNase treatment impairs dendritic cell (DC) migration from the peritoneal cavity to the draining lymph node in mice immunized i.p. with alum. However, our data show that DNase does not affect accumulation of, or expression of costimulatory proteins on, antigen-loaded DCs in lymph nodes draining injected muscles, the site by which most human vaccines are administered. DNase does inhibit prolonged T-cell-DC conjugate formation and antigen presentation between antigen-positive DCs and antigen-specific CD4 T cells following i.m. injection. Thus, from the muscle, an immunization site that does not require host DNA to promote migration of inflammatory DCs, alum acts as an adjuvant by introducing host DNA into the cytoplasm of antigen-bearing DCs, where it engages receptors that promote MHC class II presentation and better DC-T-cell interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Glenny APC, Waddington H, Wallace U. The antigenic value of toxoid precipitated by potassium alum. J Pathol Bacteriol. 1926;29:38–45.
-
- Grun JL, Maurer PH. Different T helper cell subsets elicited in mice utilizing two different adjuvant vehicles: The role of endogenous interleukin 1 in proliferative responses. Cell Immunol. 1989;121(1):134–145. - PubMed
-
- Brewer JM, et al. Aluminium hydroxide adjuvant initiates strong antigen-specific Th2 responses in the absence of IL-4- or IL-13-mediated signaling. J Immunol. 1999;163(12):6448–6454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

