Visualization of actin filaments and monomers in somatic cell nuclei
- PMID: 23447706
- PMCID: PMC3608506
- DOI: 10.1091/mbc.E12-09-0685
Visualization of actin filaments and monomers in somatic cell nuclei
Abstract
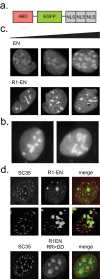
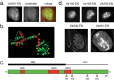
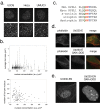
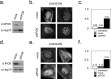
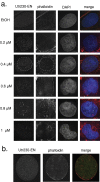
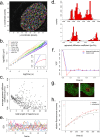
In addition to its long-studied presence in the cytoplasm, actin is also found in the nuclei of eukaryotic cells. The function and form (monomer, filament, or noncanonical oligomer) of nuclear actin are hotly debated, and its localization and dynamics are largely unknown. To determine the distribution of nuclear actin in live somatic cells and evaluate its potential functions, we constructed and validated fluorescent nuclear actin probes. Monomeric actin probes concentrate in nuclear speckles, suggesting an interaction of monomers with RNA-processing factors. Filamentous actin probes recognize discrete structures with submicron lengths that are excluded from chromatin-rich regions. In time-lapse movies, these actin filament structures exhibit one of two types of mobility: 1) diffusive, with an average diffusion coefficient of 0.06-0.08 μm(2)/s, or (2) subdiffusive, with a mobility coefficient of 0.015 μm(2)/s. Individual filament trajectories exhibit features of particles moving within a viscoelastic mesh. The small size of nuclear actin filaments is inconsistent with a role in micron-scale intranuclear transport, and their localization suggests that they do not participate directly in chromatin-based processes. Our results instead suggest that actin filaments form part of a large, viscoelastic structure in the nucleoplasm and may act as scaffolds that help organize nuclear contents.
Figures

References
-
- Bañuelos S, Saraste M, Djinovic’ Carugo K. Structural comparisons of calponin homology domains: implications for actin binding. Structure. 1998;6:1419–1431. - PubMed
-
- Bohnsack MT, Stüven T, Kuhn C, Cordes VC, Gorlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

