Glucose-methanol co-utilization in Pichia pastoris studied by metabolomics and instationary ¹³C flux analysis
- PMID: 23448228
- PMCID: PMC3626722
- DOI: 10.1186/1752-0509-7-17
Glucose-methanol co-utilization in Pichia pastoris studied by metabolomics and instationary ¹³C flux analysis
Abstract
Background: Several studies have shown that the utilization of mixed carbon feeds instead of methanol as sole carbon source is beneficial for protein production with the methylotrophic yeast Pichia pastoris. In particular, growth under mixed feed conditions appears to alleviate the metabolic burden related to stress responses triggered by protein overproduction and secretion. Yet, detailed analysis of the metabolome and fluxome under mixed carbon source metabolizing conditions are missing. To obtain a detailed flux distribution of central carbon metabolism, including the pentose phosphate pathway under methanol-glucose conditions, we have applied metabolomics and instationary ¹³C flux analysis in chemostat cultivations.
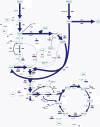
Results: Instationary ¹³C-based metabolic flux analysis using GC-MS and LC-MS measurements in time allowed for an accurate mapping of metabolic fluxes of glycolysis, pentose phosphate and methanol assimilation pathways. Compared to previous results from NMR-derived stationary state labelling data (proteinogenic amino acids, METAFoR) more fluxes could be determined with higher accuracy. Furthermore, using a thermodynamic metabolic network analysis the metabolite measurements and metabolic flux directions were validated. Notably, the concentration of several metabolites of the upper glycolysis and pentose phosphate pathway increased under glucose-methanol feeding compared to the reference glucose conditions, indicating a shift in the thermodynamic driving forces. Conversely, the extracellular concentrations of all measured metabolites were lower compared with the corresponding exometabolome of glucose-grown P. pastoris cells.The instationary ¹³C flux analysis resulted in fluxes comparable to previously obtained from NMR datasets of proteinogenic amino acids, but allowed several additional insights. Specifically, i) in vivo metabolic flux estimations were expanded to a larger metabolic network e.g. by including trehalose recycling, which accounted for about 1.5% of the glucose uptake rate; ii) the reversibility of glycolytic/gluconeogenesis, TCA cycle and pentose phosphate pathways reactions was estimated, revealing a significant gluconeogenic flux from the dihydroxyacetone phosphate/glyceraldehydes phosphate pool to glucose-6P. The origin of this finding could be carbon recycling from the methanol assimilatory pathway to the pentose phosphate pool. Additionally, high exchange fluxes of oxaloacetate with aspartate as well as malate indicated amino acid pool buffering and the activity of the malate/Asp shuttle; iii) the ratio of methanol oxidation vs utilization appeared to be lower (54 vs 79% assimilated methanol directly oxidized to CO₂).
Conclusions: In summary, the application of instationary ¹³C-based metabolic flux analysis to P. pastoris provides an experimental framework with improved capabilities to explore the regulation of the carbon and energy metabolism of this yeast, particularly for the case of methanol and multicarbon source metabolism.
Figures


Similar articles
-
Metabolic flux analysis of recombinant Pichia pastoris growing on different glycerol/methanol mixtures by iterative fitting of NMR-derived (13)C-labelling data from proteinogenic amino acids.N Biotechnol. 2014 Jan 25;31(1):120-32. doi: 10.1016/j.nbt.2013.06.007. Epub 2013 Jul 8. N Biotechnol. 2014. PMID: 23845285
-
Metabolic flux profiling of recombinant protein secreting Pichia pastoris growing on glucose:methanol mixtures.Microb Cell Fact. 2012 May 8;11:57. doi: 10.1186/1475-2859-11-57. Microb Cell Fact. 2012. PMID: 22569166 Free PMC article.
-
13 C metabolic flux profiling of Pichia pastoris grown in aerobic batch cultures on glucose revealed high relative anabolic use of TCA cycle and limited incorporation of provided precursors of branched-chain amino acids.FEBS J. 2017 Sep;284(18):3100-3113. doi: 10.1111/febs.14180. Epub 2017 Aug 7. FEBS J. 2017. PMID: 28731268
-
Pathway analysis and metabolic engineering in Corynebacterium glutamicum.Biol Chem. 2000 Sep-Oct;381(9-10):899-910. doi: 10.1515/BC.2000.111. Biol Chem. 2000. PMID: 11076021 Review.
-
Tracking the carbons supplying gluconeogenesis.J Biol Chem. 2020 Oct 16;295(42):14419-14429. doi: 10.1074/jbc.REV120.012758. Epub 2020 Aug 13. J Biol Chem. 2020. PMID: 32817317 Free PMC article. Review.
Cited by
-
Fhl1p protein, a positive transcription factor in Pichia pastoris, enhances the expression of recombinant proteins.Microb Cell Fact. 2019 Nov 29;18(1):207. doi: 10.1186/s12934-019-1256-0. Microb Cell Fact. 2019. PMID: 31783868 Free PMC article.
-
Transcriptomic Analysis of Pichia pastoris (Komagataella phaffii) GS115 During Heterologous Protein Production Using a High-Cell-Density Fed-Batch Cultivation Strategy.Front Microbiol. 2020 Mar 20;11:463. doi: 10.3389/fmicb.2020.00463. eCollection 2020. Front Microbiol. 2020. PMID: 32265887 Free PMC article.
-
Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass.Molecules. 2024 Dec 2;29(23):5695. doi: 10.3390/molecules29235695. Molecules. 2024. PMID: 39683854 Free PMC article.
-
Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris.BMC Genomics. 2022 May 12;23(1):366. doi: 10.1186/s12864-022-08592-8. BMC Genomics. 2022. PMID: 35549850 Free PMC article.
-
Identifying carbohydrate-active enzymes of Cutaneotrichosporon oleaginosus using systems biology.Microb Cell Fact. 2021 Oct 28;20(1):205. doi: 10.1186/s12934-021-01692-2. Microb Cell Fact. 2021. PMID: 34711240 Free PMC article.
References
-
- Bollok M, Resina D, Valero F, Ferrer P. Recent patents on the pichia pastoris expression system, expanding the toolbox for recombinant protein production. Recent Pat Biotechnol. 2009;3:192–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

