Metabolic biomarkers for response to PI3K inhibition in basal-like breast cancer
- PMID: 23448424
- PMCID: PMC3672699
- DOI: 10.1186/bcr3391
Metabolic biomarkers for response to PI3K inhibition in basal-like breast cancer
Abstract
Introduction: The phosphatidylinositol 3-kinase (PI3K) pathway is frequently activated in cancer cells through numerous mutations and epigenetic changes. The recent development of inhibitors targeting different components of the PI3K pathway may represent a valuable treatment alternative. However, predicting efficacy of these drugs is challenging, and methods for therapy monitoring are needed. Basal-like breast cancer (BLBC) is an aggressive breast cancer subtype, frequently associated with PI3K pathway activation. The objectives of this study were to quantify the PI3K pathway activity in tissue sections from xenografts representing basal-like and luminal-like breast cancer before and immediately after treatment with PI3K inhibitors, and to identify metabolic biomarkers for treatment response.
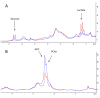
Methods: Tumor-bearing animals (n = 8 per treatment group) received MK-2206 (120 mg/kg/day) or BEZ235 (50 mg/kg/day) for 3 days. Activity in the PI3K/Akt/mammalian target of rapamycin pathway in xenografts and human biopsies was evaluated using a novel method for semiquantitative assessment of Aktser473 phosphorylation. Metabolic changes were assessed by ex vivo high-resolution magic angle spinning magnetic resonance spectroscopy.
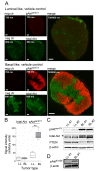
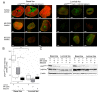
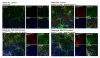
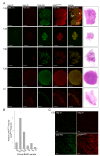
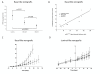
Results: Using a novel dual near-infrared immunofluorescent imaging method, basal-like xenografts had a 4.5-fold higher baseline level of pAktser473 than luminal-like xenografts. Following treatment, basal-like xenografts demonstrated reduced levels of pAktser473 and decreased proliferation. This correlated with metabolic changes, as both MK-2206 and BEZ235 reduced lactate concentration and increased phosphocholine concentration in the basal-like tumors. BEZ235 also caused increased glucose and glycerophosphocholine concentrations. No response to treatment or change in metabolic profile was seen in luminal-like xenografts. Analyzing tumor sections from five patients with BLBC demonstrated that two of these patients had an elevated pAktser473 level.
Conclusion: The activity of the PI3K pathway can be determined in tissue sections by quantitative imaging using an antibody towards pAktser473. Long-term treatment with MK-2206 or BEZ235 resulted in significant growth inhibition in basal-like, but not luminal-like, xenografts. This indicates that PI3K inhibitors may have selective efficacy in basal-like breast cancer with increased PI3K signaling, and identifies lactate, phosphocholine and glycerophosphocholine as potential metabolic biomarkers for early therapy monitoring. In human biopsies, variable pAktser473 levels were observed, suggesting heterogeneous PI3K signaling activity in BLBC.
Figures

Similar articles
-
Combined targeting of mTOR and AKT is an effective strategy for basal-like breast cancer in patient-derived xenograft models.Mol Cancer Ther. 2013 Aug;12(8):1665-75. doi: 10.1158/1535-7163.MCT-13-0159. Epub 2013 May 20. Mol Cancer Ther. 2013. PMID: 23689832 Free PMC article.
-
Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models.Breast Cancer Res. 2013 Jan 23;15(1):R8. doi: 10.1186/bcr3379. Breast Cancer Res. 2013. PMID: 23343422 Free PMC article.
-
Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells.Breast Cancer Res. 2008;10(6):R101. doi: 10.1186/bcr2204. Epub 2008 Dec 3. Breast Cancer Res. 2008. PMID: 19055754 Free PMC article.
-
Research progress and development strategy of PI3K inhibitors for breast cancer treatment: A review (2016-present).Bioorg Chem. 2024 Dec;153:107934. doi: 10.1016/j.bioorg.2024.107934. Epub 2024 Oct 28. Bioorg Chem. 2024. PMID: 39509786 Review.
-
Efficacy of PI3K inhibitors in advanced breast cancer.Ann Oncol. 2019 Dec 1;30(Suppl_10):x12-x20. doi: 10.1093/annonc/mdz381. Ann Oncol. 2019. PMID: 31859349 Free PMC article. Review.
Cited by
-
Patient-Derived Xenografts of Breast Cancer.Adv Exp Med Biol. 2025;1464:109-121. doi: 10.1007/978-3-031-70875-6_7. Adv Exp Med Biol. 2025. PMID: 39821023 Review.
-
ASSIGN: context-specific genomic profiling of multiple heterogeneous biological pathways.Bioinformatics. 2015 Jun 1;31(11):1745-53. doi: 10.1093/bioinformatics/btv031. Epub 2015 Jan 22. Bioinformatics. 2015. PMID: 25617415 Free PMC article.
-
Breast tumor subgroups reveal diverse clinical prognostic power.Sci Rep. 2014 Feb 6;4:4002. doi: 10.1038/srep04002. Sci Rep. 2014. PMID: 24499868 Free PMC article.
-
RIP1K and RIP3K provoked by shikonin induce cell cycle arrest in the triple negative breast cancer cell line, MDA-MB-468: necroptosis as a desperate programmed suicide pathway.Tumour Biol. 2016 Apr;37(4):4479-91. doi: 10.1007/s13277-015-4258-5. Epub 2015 Oct 26. Tumour Biol. 2016. PMID: 26496737
-
Proteomic characterization of breast cancer xenografts identifies early and late bevacizumab-induced responses and predicts effective drug combinations.Clin Cancer Res. 2014 Jan 15;20(2):404-12. doi: 10.1158/1078-0432.CCR-13-1865. Epub 2013 Nov 5. Clin Cancer Res. 2014. PMID: 24192926 Free PMC article.
References
-
- Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. - PubMed
-
- Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. - DOI - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

