Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways
- PMID: 23449645
- PMCID: PMC3613468
- DOI: 10.1104/pp.112.212423
Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways
Abstract
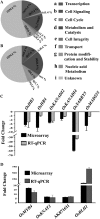
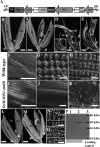
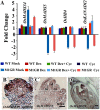
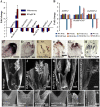
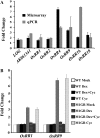
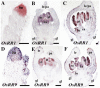
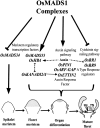
SEPALLATA (SEP) MADS box transcription factors mediate floral development in association with other regulators. Mutants in five rice (Oryza sativa) SEP genes suggest both redundant and unique functions in panicle branching and floret development. leafy hull sterile1/OsMADS1, from a grass-specific subgroup of LOFSEP genes, is required for specifying a single floret on the spikelet meristem and for floret organ development, but its downstream mechanisms are unknown. Here, key pathways and directly modulated targets of OsMADS1 were deduced from expression analysis after its knockdown and induction in developing florets and by studying its chromatin occupancy at downstream genes. The negative regulation of OsMADS34, another LOFSEP gene, and activation of OsMADS55, a SHORT VEGETATIVE PHASE-like floret meristem identity gene, show its role in facilitating the spikelet-to-floret meristem transition. Direct regulation of other transcription factor genes like OsHB4 (a class III homeodomain Leu zipper member), OsBLH1 (a BEL1-like homeodomain member), OsKANADI2, OsKANADI4, and OsETTIN2 show its role in meristem maintenance, determinacy, and lateral organ development. We found that the OsMADS1 targets OsETTIN1 and OsETTIN2 redundantly ensure carpel differentiation. The multiple effects of OsMADS1 in promoting auxin transport, signaling, and auxin-dependent expression and its direct repression of three cytokinin A-type response regulators show its role in balancing meristem growth, lateral organ differentiation, and determinacy. Overall, we show that OsMADS1 integrates transcriptional and signaling pathways to promote rice floret specification and development.
Figures

References
-
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 - PubMed
-
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 - PubMed
-
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

