The genetic architecture of degenerin/epithelial sodium channels in Drosophila
- PMID: 23449991
- PMCID: PMC3583452
- DOI: 10.1534/g3.112.005272
The genetic architecture of degenerin/epithelial sodium channels in Drosophila
Abstract
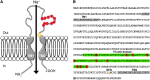
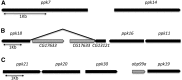
Degenerin/epithelial sodium channels (DEG/ENaC) represent a large family of animal-specific membrane proteins. Although the physiological functions of most family members are not known, some have been shown to act as nonvoltage gated, amiloride-sensitive sodium channels. The DEG/ENaC family is exceptionally large in genomes of Drosophila species relative to vertebrates and other insects. To elucidate the evolutionary history of the DEG/ENaC family in Drosophila, we took advantage of the genomic and genetic information available for 12 Drosophila species that represent all the major species groups in the Drosophila clade. We have identified 31 family members (termed pickpocket genes) in Drosophila melanogaster, which can be divided into six subfamilies, which are represented in all 12 species. Structure prediction analyses suggested that some subunits evolved unique structural features in the large extracellular domain, possibly supporting mechanosensory functions. This finding is further supported by experimental data that show that both ppk1 and ppk26 are expressed in multidendritic neurons, which can sense mechanical nociceptive stimuli in larvae. We also identified representative genes from five of the six DEG/ENaC subfamilies in a mosquito genome, suggesting that the core DEG/ENaC subfamilies were already present early in the dipteran radiation. Spatial and temporal analyses of expression patterns of the various pickpocket genes indicated that paralogous genes often show very different expression patterns, possibly indicating that gene duplication events have led to new physiological or cellular functions rather than redundancy. In summary, our analyses support a rapid early diversification of the DEG/ENaC family in Diptera followed by physiological and/or cellular specialization. Some members of the family may have diversified to support the physiological functions of a yet unknown class of ligands.
Keywords: Degenerin/epithelial sodium channel; chemosensation; fruit fly; mechanosensation; phylogeny.
Figures



Similar articles
-
Balboa binds to pickpocket in vivo and is required for mechanical nociception in Drosophila larvae.Curr Biol. 2014 Dec 15;24(24):2920-5. doi: 10.1016/j.cub.2014.10.038. Epub 2014 Nov 26. Curr Biol. 2014. PMID: 25454784 Free PMC article.
-
The Drosophila Postsynaptic DEG/ENaC Channel ppk29 Contributes to Excitatory Neurotransmission.J Neurosci. 2017 Mar 22;37(12):3171-3180. doi: 10.1523/JNEUROSCI.3850-16.2017. Epub 2017 Feb 17. J Neurosci. 2017. PMID: 28213447 Free PMC article.
-
Identification of Ppk26, a DEG/ENaC Channel Functioning with Ppk1 in a Mutually Dependent Manner to Guide Locomotion Behavior in Drosophila.Cell Rep. 2014 Nov 20;9(4):1446-58. doi: 10.1016/j.celrep.2014.10.034. Epub 2014 Nov 6. Cell Rep. 2014. PMID: 25456135 Free PMC article.
-
Sensory functions for degenerin/epithelial sodium channels (DEG/ENaC).Adv Genet. 2011;76:1-26. doi: 10.1016/B978-0-12-386481-9.00001-8. Adv Genet. 2011. PMID: 22099690 Free PMC article. Review.
-
Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure.Physiol Rev. 2002 Jul;82(3):735-67. doi: 10.1152/physrev.00007.2002. Physiol Rev. 2002. PMID: 12087134 Review.
Cited by
-
Genomic architecture and functional unit of mimicry supergene in female limited Batesian mimic Papilio butterflies.Philos Trans R Soc Lond B Biol Sci. 2022 Aug;377(1856):20210198. doi: 10.1098/rstb.2021.0198. Epub 2022 Jun 13. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35694751 Free PMC article. Review.
-
The neural basis for insect pheromonal communication.Curr Opin Insect Sci. 2015 Dec 1;12:86-92. doi: 10.1016/j.cois.2015.09.010. Epub 2015 Oct 24. Curr Opin Insect Sci. 2015. PMID: 26568912 Free PMC article.
-
CG4928 Is Vital for Renal Function in Fruit Flies and Membrane Potential in Cells: A First In-Depth Characterization of the Putative Solute Carrier UNC93A.Front Cell Dev Biol. 2020 Oct 14;8:580291. doi: 10.3389/fcell.2020.580291. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33163493 Free PMC article.
-
Mosquito Cell Atlas: A single-nucleus transcriptomic atlas of the adult Aedes aegypti mosquito.bioRxiv [Preprint]. 2025 Feb 25:2025.02.25.639765. doi: 10.1101/2025.02.25.639765. bioRxiv. 2025. PMID: 40060408 Free PMC article. Preprint.
-
Bioelectric regulation of intestinal stem cells.Trends Cell Biol. 2023 Jul;33(7):555-567. doi: 10.1016/j.tcb.2022.10.003. Epub 2022 Nov 15. Trends Cell Biol. 2023. PMID: 36396487 Free PMC article. Review.
References
-
- Abascal F., Zardoya R., Posada D., 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 - PubMed
-
- Adams C. M., Snyder P. M., Welsh M. J., 1997. Interactions between subunits of the human epithelial sodium channel. J. Biol. Chem. 272: 27295–27300 - PubMed
-
- Ainsley J. A., Pettus J. M., Bosenko D., Gerstein C. E., Zinkevich N., et al. , 2003. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 13: 1557–1563 - PubMed
-
- Arnadottir J., Chalfie M., 2010. Eukaryotic mechanosensitive channels. Annu Rev Biophys 39: 111–137 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
