A genomic survey of Reb homologs suggests widespread occurrence of R-bodies in proteobacteria
- PMID: 23450193
- PMCID: PMC3583457
- DOI: 10.1534/g3.112.005231
A genomic survey of Reb homologs suggests widespread occurrence of R-bodies in proteobacteria
Abstract
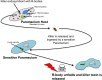
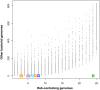
Bacteria and eukaryotes are involved in many types of interaction in nature, with important ecological consequences. However, the diversity, occurrence, and mechanisms of these interactions often are not fully known. The obligate bacterial endosymbionts of Paramecium provide their hosts with the ability to kill sensitive Paramecium strains through the production of R-bodies, highly insoluble coiled protein ribbons. R-bodies have been observed in a number of free-living bacteria, where their function is unknown. We have performed an exhaustive survey of genes coding for homologs of Reb proteins (R-body components) in complete bacterial genomes. We found that reb genes are much more widespread than previously thought, being present in representatives of major Proteobacterial subdivisions, including many free-living taxa, as well as taxa known to be involved in various kinds of interactions with eukaryotes, from mutualistic associations to pathogenicity. Reb proteins display very good conservation at the sequence level, suggesting that they may produce functional R-bodies. Phylogenomic analysis indicates that reb genes underwent a complex evolutionary history and allowed the identification of candidates potentially involved in R-body assembly, functioning, regulation, or toxicity. Our results strongly suggest that the ability to produce R-bodies is likely widespread in Proteobacteria. The potential involvement of R-bodies in as yet unexplored interactions with eukaryotes and the consequent ecological implications are discussed.
Keywords: Caedibacter; kappa particles; phylogenomics.
Figures





References
-
- Beale G., Preer J. R., Jr, 2004. Paramecium: Genetics and Epigenetics, CRC Press, Boca Raton, FL
-
- Brown M. R., Barker J., 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7: 46–50 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
