On the mutational topology of the bacterial genome
- PMID: 23450823
- PMCID: PMC3583449
- DOI: 10.1534/g3.112.005355
On the mutational topology of the bacterial genome
Abstract
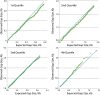
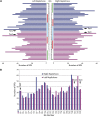
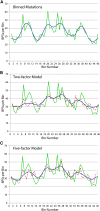
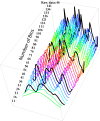
By sequencing the genomes of 34 mutation accumulation lines of a mismatch-repair defective strain of Escherichia coli that had undergone a total of 12,750 generations, we identified 1625 spontaneous base-pair substitutions spread across the E. coli genome. These mutations are not distributed at random but, instead, fall into a wave-like spatial pattern that is repeated almost exactly in mirror image in the two separately replicated halves of the bacterial chromosome. The pattern is correlated to genomic features, with mutation densities greatest in regions predicted to have high superhelicity. Superimposed upon this pattern are regional hotspots, some of which are located where replication forks may collide or be blocked. These results suggest that, as they traverse the chromosome, the two replication forks encounter parallel structural features that change the fidelity of DNA replication.
Keywords: DNA polymerase errors; chromosome structure; evolution; mutation rate; replication fidelity.
Figures

References
-
- Agier N., Fischer G., 2012. The mutational profile of the yeast genome is shaped by replication. Mol. Biol. Evol. 29: 905–913 - PubMed
-
- Akaike H., 1973. Information theory and the extension of the maximum likelihood principle, pp. 267–281 in Second International Symposium on Information Theory, edited by Petrov V. N., Csaki F. Academiai Kiadó, Budapest
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
