Systemic amyloidoses
- PMID: 23451869
- PMCID: PMC4044913
- DOI: 10.1146/annurev-biochem-072611-130030
Systemic amyloidoses
Abstract
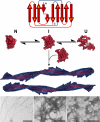
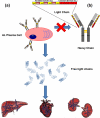
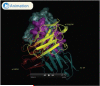
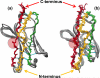
The amyloidoses are a group of protein misfolding diseases in which the precursor protein undergoes a conformational change that triggers the formation of amyloid fibrils in different tissues and organs, causing cell death and organ failure. Amyloidoses can be either localized or systemic. In localized amyloidosis, amyloid deposits form at the site of precursor protein synthesis, whereas in systemic amyloidosis, amyloid deposition occurs distant from the site of precursor protein secretion. We review the type of proteins and cells involved and what is known about the complex pathophysiology of these diseases. We focus on light chain amyloidosis to illustrate how biochemical and biophysical studies have led to a deeper understanding of the pathogenesis of this devastating disease. We also review current cellular, tissue, and animal models and discuss the challenges and opportunities for future studies of the systemic amyloidoses.
Figures

References
-
- Sipe JD, Benson MD, Buxbaum JN, Ikeda S-I, Merlini G, et al. Amyloid fibril protein nomenclature: 2010 recommendations from the nomenclature committee of the International Society of Amyloidosis. Amyloid. 2010;17:101–04. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. - PubMed
-
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–39. - PubMed
-
- Geddes AJ, Parker KD, Atkins EDT, Beighton E. “Cross-β” conformation in proteins. Journal of Molecular Biology. 1968;32:343–58. - PubMed
-
- Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. Journal of Histochemistry & Cytochemistry. 1968;16:673–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

