Influences of membrane mimetic environments on membrane protein structures
- PMID: 23451886
- PMCID: PMC3731949
- DOI: 10.1146/annurev-biophys-083012-130326
Influences of membrane mimetic environments on membrane protein structures
Abstract
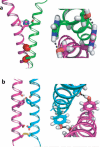
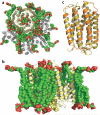
The number of membrane protein structures in the Protein Data Bank is becoming significant and growing. Here, the transmembrane domain structures of the helical membrane proteins are evaluated to assess the influences of the membrane mimetic environments. Toward this goal, many of the biophysical properties of membranes are discussed and contrasted with those of the membrane mimetics commonly used for structure determination. Although the mimetic environments can perturb the protein structures to an extent that potentially gives rise to misinterpretation of functional mechanisms, there are also many structures that have a native-like appearance. From this assessment, an initial set of guidelines is proposed for distinguishing native-like from nonnative-like membrane protein structures. With experimental techniques for validation and computational methods for refinement and quality assessment and enhancement, there are good prospects for achieving native-like structures for these very important proteins.
Figures
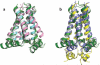
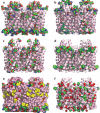

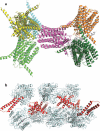

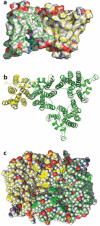

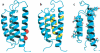
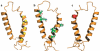
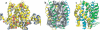
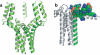
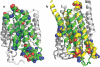
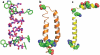
References
-
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. - PubMed
-
- Belrhali H, Nollert P, Royant A, Menzel C, Rosenbusch JP, et al. Protein, lipid water organization in bacteriorhodopsin: a molecular view of the purple membrane at 1.9Å resolution. Structure. 1999;7:909–17. - PubMed
-
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

