Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation
- PMID: 23451887
- PMCID: PMC3737589
- DOI: 10.1146/annurev-biophys-083012-130358
Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation
Abstract
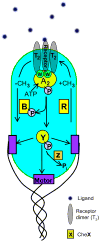
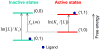
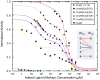
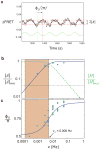
We review the recent developments in understanding the bacterial chemotaxis signaling pathway by using quantitative modeling methods. The models developed are based on structural information of the signaling complex and the dynamics of the underlying biochemical network. We focus on two important functions of the bacterial chemotaxis signaling pathway: signal amplification and adaptation. We describe in detail the structure and the dynamics of the mathematical models and how they compare with existing experiments, emphasizing the predictability of the models. Finally, we outline future directions for developing the modeling approach to better understand the bacterial chemosensory system.
Figures




References
-
- Adler J, Tso WW. Desicion-making in bacteria: chemotactic response of E. coli to conflicting stimuli. Science. 1976;184:1292–1294. - PubMed
-
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in bachterial chemotaxis. Nature. 1999;397:168–171. - PubMed
-
- Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. First model to explain accurate robust adaptation in E. coli chemo-taxis. - PubMed
-
- Berg HC, Brown DA. Chemotaxis in E. coli analysed by Three dimensional Tracking. Nature. 1972;239(5374):500–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

