Profiling of ubiquitin-like modifications reveals features of mitotic control
- PMID: 23452859
- PMCID: PMC3711129
- DOI: 10.1016/j.cell.2013.02.007
Profiling of ubiquitin-like modifications reveals features of mitotic control
Abstract
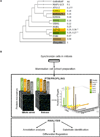
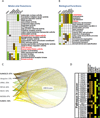
Ubiquitin and ubiquitin-like (Ubl) protein modifications affect protein stability, activity, and localization, but we still lack broad understanding of the functions of Ubl modifications. We have profiled the protein targets of ubiquitin and six additional Ubls in mitosis using a functional assay that utilizes active mammalian cell extracts and protein microarrays and identified 1,500 potential substrates; 80-200 protein targets were exclusive to each Ubl. The network structure is nonrandom, with most targets mapping to a single Ubl. There are distinct molecular functions for each Ubl, suggesting divergent biological roles. Analysis of differential profiles between mitosis and G1 highlighted a previously underappreciated role for the Ubl, FAT10, in mitotic regulation. In addition to its role as a resource for Ubl modifications, our study provides a systematic approach to analyze changes in posttranslational modifications at various cellular states.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Aichem A, Kalveram B, Spinnenhirn V, Kluge K, Catone N, Johansen T, Groettrup M. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci. 2012;125:4576–4585. - PubMed
-
- Aichem A, Pelzer C, Lukasiak S, Kalveram B, Sheppard PW, Rani N, Schmidtke G, Groettrup M. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010a;1:1–10. - PubMed
-
- Aichem A, Pelzer C, Lukasiak S, Kalveram B, Sheppard PW, Rani N, Schmidtke G, Groettrup M. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Commun. 2010b;1:13. - PubMed
-
- Ban R, Nishida T, Urano T. Mitotic kinase Aurora-B is regulated by SUMO-2/3 conjugation/deconjugation during mitosis. Genes Cells. 2011;16:652–669. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

