N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function
- PMID: 23453015
- PMCID: PMC4357660
- DOI: 10.1016/j.gpb.2012.12.002
N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function
Abstract
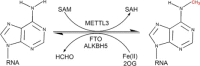
N(6)-methyl-adenosine (m(6)A) is one of the most common and abundant modifications on RNA molecules present in eukaryotes. However, the biological significance of m(6)A methylation remains largely unknown. Several independent lines of evidence suggest that the dynamic regulation of m(6)A may have a profound impact on gene expression regulation. The m(6)A modification is catalyzed by an unidentified methyltransferase complex containing at least one subunit methyltransferase like 3 (METTL3). m(6)A modification on messenger RNAs (mRNAs) mainly occurs in the exonic regions and 3'-untranslated region (3'-UTR) as revealed by high-throughput m(6)A-seq. One significant advance in m(6)A research is the recent discovery of the first two m(6)A RNA demethylases fat mass and obesity-associated (FTO) gene and ALKBH5, which catalyze m(6)A demethylation in an α-ketoglutarate (α-KG)- and Fe(2+)-dependent manner. Recent studies in model organisms demonstrate that METTL3, FTO and ALKBH5 play important roles in many biological processes, ranging from development and metabolism to fertility. Moreover, perturbation of activities of these enzymes leads to the disturbed expression of thousands of genes at the cellular level, implicating a regulatory role of m(6)A in RNA metabolism. Given the vital roles of DNA and histone methylations in epigenetic regulation of basic life processes in mammals, the dynamic and reversible chemical m(6)A modification on RNA may also serve as a novel epigenetic marker of profound biological significances.
Copyright © 2013. Production and hosting by Elsevier Ltd.
Figures



References
-
- Narayan P., Rottman F.M. Methylation of mRNA. Adv Enzymol Relat Areas Mol Biol. 1992;65:255–285. - PubMed
-
- Kennedy T.D., Lane B.G. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5′-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Can J Biochem. 1979;57:927–931. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

