Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells
- PMID: 23453631
- PMCID: PMC3634355
- DOI: 10.1016/j.immuni.2013.02.010
Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells
Abstract
Mucosal innate lymphoid cell (ILC) subsets promote immune responses to pathogens by producing distinct signature cytokines in response to changes in the cytokine microenvironment. We previously identified human ILC3 distinguished by interleukin-22 (IL-22) secretion. Here we characterized a human ILC1 subset that produced interferon-γ (IFN-γ) in response to IL-12 and IL-15 and had a unique integrin profile, intraepithelial location, hallmarks of TGF-β imprinting, and a memory-activated phenotype. Because tissue-resident memory CD8(+) T cells share this profile, intraepithelial ILC1 may be their innate counterparts. In mice, intraepithelial ILC1 were distinguished by CD160 expression and required Nfil3- and Tbx21-encoded transcription factors for development, but not IL-15 receptor-α, indicating that intraepithelial ILC1 are distinct from conventional NK cells. Intraepithelial ILC1 were amplified in Crohn's disease patients and contributed to pathology in the anti-CD40-induced colitis model in mice. Thus, intraepithelial ILC1 may initiate IFN-γ responses against pathogens but contribute to pathology when dysregulated.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

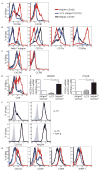
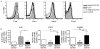
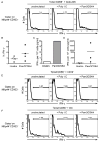
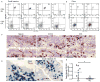


Comment in
-
ILC1 populations join the border patrol.Immunity. 2013 Apr 18;38(4):630-2. doi: 10.1016/j.immuni.2013.03.005. Immunity. 2013. PMID: 23601681
References
-
- Allez M, Brimnes J, Dotan I, Mayer L. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology. 2002;123:1516–1526. - PubMed
-
- Carman PS, Ernst PB, Rosenthal KL, Clark DA, Befus AD, Bienenstock J. Intraepithelial leukocytes contain a unique subpopulation of NK-like cytotoxic cells active in the defense of gut epithelium to enteric murine coronavirus. J Immunol. 1986;136:1548–1553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

